Nuclear retention of unspliced HIV-1 RNA as a reversible post-transcriptional block in latency
- PMID: 40021667
- PMCID: PMC11871326
- DOI: 10.1038/s41467-025-57290-y
Nuclear retention of unspliced HIV-1 RNA as a reversible post-transcriptional block in latency
Abstract
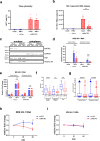
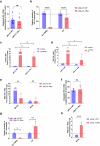
HIV-1 latency is mainly characterized at transcriptional level, and little is known about post-transcriptional mechanisms and their contribution to reactivation. The viral protein Rev controls the nucleocytoplasmic export of unspliced and singly-spliced RNA that is central to proviral replication-competence and is therefore a prerequisite for efficient viral reactivation during the "shock-and-kill" cure therapy. Here we show that during infection and reactivation, unspliced HIV-1 RNA is a subject to complex and dynamic regulation by the Rev cofactor MATR3 and the MTR4 cofactor of the nuclear exosome. MATR3 and MTR4 coexist in the same ribonucleoprotein complex functioning to either maintain or degrade the RNA, respectively, with Rev orchestrating this regulatory switch. Moreover, we provide evidence of nuclear retention of unspliced HIV-1 RNA in ex vivo cultures from 22 ART-treated people with HIV, highlighting a reversible post-transcriptional block to viral RNA nucleocytoplasmic export that is relevant to the design of curative interventions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.O.P. received a research grant from Gilead Sciences Research Program. C.V.L. received a research grant from ViiV Healthcare. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The remaining authors declare no competing interests.
Figures


References
-
- Pasternak, A. O. & Berkhout, B. HIV persistence: silence or resistance? Curr. Opin. Virol.59, 101301 (2023). - PubMed
-
- Crowe, S., Zhu, T. & Muller, W. A. The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. J. Leukoc. Biol.74, 635–641 (2003). - PubMed
-
- Lewin, S. R. et al. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS12, 719–727 (1998). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

