Enterocloster clostridioformis protects against Salmonella pathogenesis and modulates epithelial and mucosal immune function
- PMID: 40022210
- PMCID: PMC11869688
- DOI: 10.1186/s40168-025-02050-9
Enterocloster clostridioformis protects against Salmonella pathogenesis and modulates epithelial and mucosal immune function
Abstract
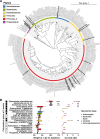
Background: Promoting resistance to enteric pathogen infection is a core function of the gut microbiota; however, many of the specific host-commensal interactions that mediate this protection remain uncharacterised. To address this knowledge gap, we monocolonised germ-free mice with mouse-derived commensal microbes to screen for microbiota-induced resistance to Salmonella Typhimurium infection.
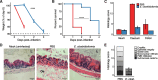
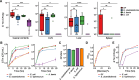
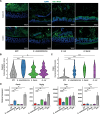
Results: We identified Enterocloster clostridioformis as a protective species against S. Typhimurium infection. E. clostridioformis selectively upregulates resistin-like molecule β and cell cycle pathway expression at the level of caecal epithelial cells and increases T-regulatory cells in the underlying mucosal immune system, potentially contributing to reduced infection-induced pathology.
Conclusions: We highlight novel mechanisms of host-microbe interactions that can mediate microbiota-induced resistance to acute salmonellosis. In the backdrop of increasing antibiotic resistance, this study identifies novel potential avenues for further research into protective host responses against enteric infections and could lead to new therapeutic approaches. Video Abstract.
Keywords: Enterocloster clostridioformis; Escherichia coli; Acute salmonellosis; Host–commensal interactions; Microbiota; Mouse models; Pathogen resistance; Resistin-like molecule β; Screen; T-regulatory cells.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Mice were maintained at the Home Office-approved Wellcome Sanger Institute mouse facility, and all procedures were carried out in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986. Consent for publication: Not applicable. Competing interests: TDL is a founder and CSO of Microbiotica. The other authors declare that they have no competing interests.
Figures

References
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease 'Burden of Illness S. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882–9. - PubMed
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;12(12):e1001921. - PMC - PubMed
-
- Tasmin R, Gulig PA, Parveen S. Detection of virulence plasmid-encoded genes in Salmonella Typhimurium and Salmonella Kentucky isolates recovered from commercially processed chicken carcasses. J Food Prot. 2019;82(8):1364–8. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

