HIF regulates multiple translated endogenous retroviruses: Implications for cancer immunotherapy
- PMID: 40023154
- PMCID: PMC11988688
- DOI: 10.1016/j.cell.2025.01.046
HIF regulates multiple translated endogenous retroviruses: Implications for cancer immunotherapy
Abstract
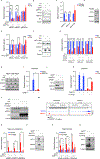
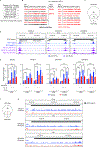
Clear cell renal cell carcinoma (ccRCC), despite having a low mutational burden, is considered immunogenic because it occasionally undergoes spontaneous regressions and often responds to immunotherapies. The signature lesion in ccRCC is inactivation of the VHL tumor suppressor gene and consequent upregulation of the HIF transcription factor. An earlier case report described a ccRCC patient who was cured by an allogeneic stem cell transplant and later found to have donor-derived T cells that recognized a ccRCC-specific peptide encoded by a HIF-responsive endogenous retrovirus (ERV), ERVE-4. We report that ERVE-4 is one of many ERVs that are induced by HIF, translated into HLA-bound peptides in ccRCCs, and capable of generating antigen-specific T cell responses. Moreover, ERV expression can be induced in non-ccRCC tumors with clinical-grade HIF stabilizers. These findings have implications for leveraging ERVs for cancer immunotherapy.
Keywords: ERV; HIF; cancer vaccine; ccRCC; immunopeptidomic; immunotherapy; kidney cancer; neoantigen.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.A.B. reports share options in Elephas; advisory board, consulting, or personal fees from DLA Piper, AbbVie, Compugen, Link Cell Therapies, Scholar Rock, NeoMorph, Nimbus, Exelixis, AVEO, Eisai, and Elephas; and research support from AstraZeneca, outside of the submitted work. N.N., T.P., M.C.-R., C.G., and G.M. are employees of and own equity in T-Scan Therapeutics, and G.M. is a board member of T-Scan Therapeutics. S.K. is currently an employee of Genentech. W.J.L. reports advisory board and consulting fees from One Lambda Inc. and CareDx Inc. and royalties from Thermo Fisher Scientific Inc. S.A.S. is a consultant for Jivanu Therapeutics and has equity in Agenus Inc., Agios Pharmaceuticals, Breakbio Corp., Bristol-Myers Squibb, and Lumos Pharma. E.M.V.A. provides advisory/consulting to Enara Bio, Manifold Bio, Monte Rosa, and the Novartis Institute for Biomedical Research; receives research support from Novartis, BMS, Sanofi, and NextPoint; and owns equity in Tango Therapeutics, Enara Bio, and Monte Rosa. T.K.C. reports involvement in NeoVax clinical trials and Gateway for Cancer Research; advisory board, consulting, or personal fees from Merck and BMS; institutional patents (outside the submitted work) filed on molecular alterations and immunotherapy response/toxicity and ctDNA; and equity in Tempest, Osel, Precede Bio, CureResponse, InnDura Therapeutics, Primium, Abalytics, and Faron Pharma. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, Seer, and PrognomIQ. C.J.W. holds equity at BioNTech, Inc. W.G.K. has financial interests in Lilly Pharmaceuticals, Fibrogen, Nextech Invest, and Casdin Capital and receives belzutifan royalties.
Figures





References
-
- Sharma P, Siddiqui BA, Anandhan S, Yadav SS, Subudhi SK, Gao J, Goswami S, and Allison JP (2021). The Next Decade of Immune Checkpoint Therapy. Cancer Discov 11, 838–857. 10.1158/2159-8290.CD-20-1680. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- U54 CA272688/CA/NCI NIH HHS/United States
- P50 CA101942/CA/NCI NIH HHS/United States
- R35 CA210068/CA/NCI NIH HHS/United States
- R01 CA285308/CA/NCI NIH HHS/United States
- R01 HL157174/HL/NHLBI NIH HHS/United States
- P01 CA206978/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 CA155010/CA/NCI NIH HHS/United States
- R01 GM123002/GM/NIGMS NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- R01 CA279391/CA/NCI NIH HHS/United States
- U01 CA271402/CA/NCI NIH HHS/United States
- U24 CA270823/CA/NCI NIH HHS/United States
- T32 CA009172/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

