Upregulation of the interferon-inducible antiviral gene RSAD2 in neuroendocrine prostate cancer via PVT1 exon 9 dependent and independent pathways
- PMID: 40024473
- PMCID: PMC11994405
- DOI: 10.1016/j.jbc.2025.108370
Upregulation of the interferon-inducible antiviral gene RSAD2 in neuroendocrine prostate cancer via PVT1 exon 9 dependent and independent pathways
Abstract
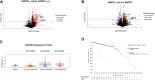
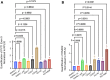
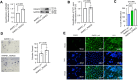
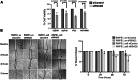
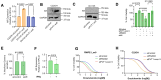
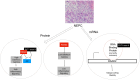
PVT1 exon 9 overexpression is a newly uncovered aberration in prostate cancer (PCa). We have previously demonstrated the exon 9 region of PVT1 is overexpressed in some patient PCa tissues and caused development of neuroendocrine prostate cancer (NEPC) in vitro and in vivo. In this study, we focused on elucidating downstream mechanisms induced by PVT1 exon 9 overexpression with the goal of further understanding its role in NEPC development. RNA-seq analysis of a PVT1 exon 9 overexpressing PCa model revealed significant enrichment of genes responsible for inducing inflammatory processes including RSAD2. We observed RSAD2 overexpression in all NEPC models examined whereas PVT1 exon 9 was overexpressed only in a subset of the NEPC models. We identified two distinct pathways in which RSAD2 is overexpressed: one dependent and one independent on PVT1 exon 9 overexpression. Knockdown of RSAD2 suppressed cell proliferation and migration suggestive of its role as a therapeutic target in NEPC. We identified RSAD2 induces increased cell proliferation, colony formation, and may be involved in the transition between CRPC and NEPC. Distinct differences between PVT1 exon 9-dependent and PVT1 exon 9-independent NEPC models include differences in type II interferon signaling and AR modulation. PVT1 exon 9 binds to RSAD2 protein and disruption of binding significantly impedes downstream interferon gamma secretion by PVT1 exon 9-dependent NEPC cells. These novel findings indicate the importance of these two independent pathways in NEPC, the need to identify relevant NEPC patient populations and study strategies for targeting PVT1 exon 9 and/or RSAD2.
Keywords: PVT1; androgen receptor; cell biology; cellular immune response; interferon; long-noncoding RNA; neuroendocrine; prostate cancer.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest O. O. O. is the co-founder of NucleoBio Inc, a start-up biotechnology company. All other authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

