Chenodeoxycholic acid modulates cholestatic niche through FXR/Myc/P-selectin axis in liver endothelial cells
- PMID: 40025016
- PMCID: PMC11873286
- DOI: 10.1038/s41467-025-57351-2
Chenodeoxycholic acid modulates cholestatic niche through FXR/Myc/P-selectin axis in liver endothelial cells
Abstract
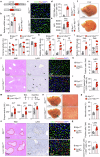
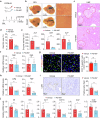
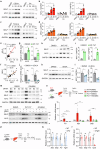
Cholestatic liver diseases are characterized by excessive bile acid accumulation in the liver. Endothelial cells (ECs) shape the local microenvironment in both normal conditions and liver injury, yet their role in cholestasis is unclear. Through a comparative analysis of single-cell RNA sequencing data from various murine models of liver injury, we identify distinctive Myc activation within ECs during obstructive cholestasis resulting from bile duct ligation (BDL). Myc overexpression in ECs significantly upregulates P-selectin, increasing neutrophil infiltration and worsening cholestatic liver injury. This process occurs through the FXR, activated by chenodeoxycholic acid (CDCA) and its conjugate TCDCA. Inhibiting P-selectin with PSI-697 reduces neutrophil recruitment and alleviates injury. Cholestatic patient liver samples also show elevated Myc and P-selectin in ECs, along with increased neutrophils. The findings identify ECs as key drivers of cholestatic liver injury through a Myc-driven program and suggest that targeting the CDCA/FXR/Myc/P-selectin axis may offer a therapeutic approach.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Hirschfield, G. M., Heathcote, E. J. & Gershwin, M. E. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology139, 1481–1496 (2010). - PubMed
-
- Guo, C. S. et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity45, 802–816 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- 82071854/National Natural Science Foundation of China (National Science Foundation of China)
- 32100590/National Natural Science Foundation of China (National Science Foundation of China)
- 32270820/National Natural Science Foundation of China (National Science Foundation of China)
- 32422024/National Natural Science Foundation of China (National Science Foundation of China)
- ZR2020QH038/Natural Science Foundation of Shandong Province (Shandong Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources

