Cellular immune breadth of an Omicron-specific, self-amplifying monovalent mRNA vaccine booster for COVID-19
- PMID: 40025095
- PMCID: PMC11873296
- DOI: 10.1038/s41541-025-01076-2
Cellular immune breadth of an Omicron-specific, self-amplifying monovalent mRNA vaccine booster for COVID-19
Abstract
Selecting a booster vaccine strategy that generates cellular immune breadth is crucial for effectively recalling cellular reservoirs upon infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants. This post hoc analysis from a multicentre, randomized phase 3 study (CTRI/2022/10/046475) compared the cellular immune breadth induced by self-replicating mRNA (samRNA) vaccine GEMCOVAC-OM, encoding Omicron B.1.1.529 Spike protein, with the adenovector vaccine ChAdOx1 nCoV-19, encoding Wuhan variant Spike protein, when administered as a booster. GEMCOVAC-OM elicited significant expansion of memory B-cells (MBCs) specific to Omicron B.1.1.529, compared to ChAdOx1 nCoV-19. GEMCOVAC-OM also induced more B-cells reactive to Omicron XBB.1.5 and BA.2.86 Spike proteins. Additionally, GEMCOVAC-OM triggered higher frequencies of Omicron-Spike-specific T-cells, including stem cell, central, and effector memory subsets. In summary, while ChAdOx1 nCoV-19 showed some cross-reactivity, GEMCOVAC-OM induced a more targeted immune response. GEMCOVAC-OM offers a broader, longer-lasting immunity, making it a promising candidate for future vaccine development and global distribution.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no duality of interest associated with this manuscript. All authors were supported by the internal organization’s salary structure. Gennova Biopharmaceuticals Limited. Dr Himanshu Shashikant Pophale, Dr Prakash Sadashiv Shende, Dr Ravindra Shinde, Dr Vikram B Vikhe, Dr Abhishek Madhav Karmalkar, Dr Bhaskar Vilasrao Jedhe-Deshmukh, Dr Krishna Madhukar Giri, Dr Shrikant Vishnu Deshpande, Dr Ajay Abhimanyu Bulle, Dr Mohammed Sabah Siddiqui, Dr Swapnav Borthakur, Dr V Reddy Tummuru, Dr A. Venkateshwar Rao, Dr Dhaiwat Murugesh Shukla, Dr Manish Kumar Jain, Dr Pankaj Bhardwaj, Dr Pravin Dinkar Supe, Dr Manoj K Das, Dr Vijay Kumar Bhagwan Barge, and Dr Manoj Lahoti were clinical trial site investigators who were funded by Gennova Biopharmaceuticals Limited for their contribution to this study.
Figures
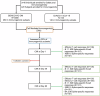


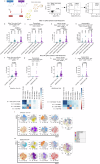

References
-
- Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol.21, 361–379 (2023). - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

