Systematic biotechnological production of isoprenoid analogs with bespoke carbon skeletons
- PMID: 40025103
- PMCID: PMC11873216
- DOI: 10.1038/s41467-025-57494-2
Systematic biotechnological production of isoprenoid analogs with bespoke carbon skeletons
Abstract
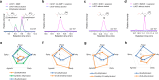
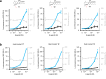
Natural products are widely used as pharmaceuticals, flavors, fragrances, and cosmetic ingredients. Synthesizing and evaluating analogs of natural products can considerably expand their applications. However, the chemical synthesis of analogs of natural products is severely hampered by their highly complex structures. This is particularly evident in isoprenoids, the largest class of natural products. Here, we develop a yeast cell-based biocatalytic method that enables the systematic biotechnological production of analogs of different classes of isoprenoids (including monoterpenoids, sesquiterpenoids, triterpenoids, and cannabinoids) with additional carbons in their skeletons. We demonstrate the applicability of this approach through two proof-of-concept studies: the biosynthesis of the highly valued aroma ingredient ethyllinalool, and the production of cannabinoid analogs with improved cannabinoid receptor agonism. This method is simple, readily adaptable to any cell factory, and enables the tailored expansion of the isoprenoid chemical space to identify molecules with improved properties and the biotechnological production of valuable compounds.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: L.W., M.R., and S.C.K. are co-inventors of a patent application (WO2023006699A1) describing the use of AtFKI to phosphorylate a primary alcohol to a mono- or pyrophosphate terpenoid precursor. A.F. is an employee of EvodiaBio ApS. S.C.K. has financial interests in EvodiaBio ApS. The remaining authors declare no competing interest.
Figures




References
-
- Zeng, T. et al. TeroKit: A database-driven web server for terpenome research. J. Chem. Inf. Model.60, 2082–2090 (2020). - PubMed
-
- Paddon, C. J. & Keasling, J. D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol.12, 355–367 (2014). - PubMed
-
- Luo, X. et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature567, 123–126 (2019). - PubMed
-
- Paddon, C. J. et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature496, 528–532 (2013). - PubMed
-
- Jiang, B. et al. Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science383, 622–629 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

