Cell-type-specific requirement for TYK2 in murine immune cells under steady state and challenged conditions
- PMID: 40025196
- PMCID: PMC11872851
- DOI: 10.1007/s00018-025-05625-9
Cell-type-specific requirement for TYK2 in murine immune cells under steady state and challenged conditions
Abstract
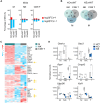
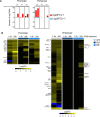
Tyrosine kinase 2 (TYK2) deficiency and loss or inhibition of kinase activity in men and mice leads to similar immune compromised phenotypes, predominantly through impairment of interferon (IFN) and interleukin 12 family responses. Here we relate the transcriptome changes to phenotypical changes observed in TYK2-deficient (Tyk2-/-) and TYK2 kinase-inactive (Tyk2K923E) mice in naïve splenic immune cells and upon ex vivo IFN treatment or in vivo tumor transplant infiltration. The TYK2 activities under homeostatic and both challenged conditions are highly cell-type-specific with respect to quantity and quality of transcriptionally dependent genes. The major impact of loss of TYK2 protein or kinase activity in splenic homeostatic macrophages, NK and CD8+ T cells and tumor-derived cytolytic cells is on IFN responses. While reportedly TYK2 deficiency leads to partial impairment of IFN-I responses, we identified cell-type-specific IFN-I-repressed gene sets completely dependent on TYK2 kinase activity. Reported kinase-inactive functions of TYK2 relate to signaling crosstalk, metabolic functions and cell differentiation or maturation. None of these phenotypes relates to respective enriched gene sets in the TYK2 kinase-inactive cell types. Nonetheless, the scaffolding functions of TYK2 are capable to change transcriptional activities at single gene levels and chromatin accessibility at promoter-distal regions upon cytokine treatment most prominently in CD8+ T cells. The cell-type-specific transcriptomic and epigenetic effects of TYK2 shed new light on the biology of this JAK family member and are relevant for current and future treatment of autoimmune and inflammatory diseases with TYK2 inhibitors.
Keywords: CD8+ T cells; Interferon; JAK-STAT; Macrophages; NK cells; Splenic immune cells; Tonic signaling; Tumor-infiltrating cells.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interests: The authors have no relevant financial or non-financial interests to disclose. Ethical Approval: All mice used in this study were bred at the University of Veterinary Medicine Vienna under specific-pathogen-free conditions according to Federation of European Laboratory Animal Science Associations (FELASA) guidelines. All experiments were performed in accordance with the European Union 2010/63 EU directive and the Austrian Federal Ministry of Science and Research according to §§ 26ff animal experimentation law. All animal experiments were approved by the Ethics and Animal Welfare Committee of the University of Veterinary Medicine Vienna Animal Experiments Act, Tierversuchsgesetz 2012—TVG 2012 (BMBWF_GZ:2020–0.200.397, approval 27.03.2020; BMWFW-68.205/0068-WF/V/3b/2015, approval 17.04.2015; BMWFW-68.205/0103-WF/V/3b/2015, approval 19.06.2015; BMBWF-68.205/0174-V/3b/2018, approval 09.10.2018) and conform to the guidelines of FELASA and ARRIVE (Animal Research: Reporting of In Vivo Experiments). Consent to Participate: Not applicable. Consent to Publish: All authors reviewed their respective results contributions and contributed to the final manuscript. All authors approved this manuscript for publication.
Figures




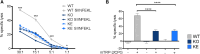


References
-
- Boisson-Dupuis S, Ramirez-Alejo N, Li Z, Patin E, Rao G, Kerner G, Lim CK, Krementsov DN, Hernandez N, Ma CS, Zhang Q, Markle J, Martinez-Barricarte R, Payne K, Fisch R, Deswarte C, Halpern J, Bouaziz M, Mulwa J, Sivanesan D, Lazarov T, Naves R, Garcia P, Itan Y, Boisson B, Checchi A, Jabot-Hanin F, Cobat A, Guennoun A, Jackson CC, Pekcan S, Caliskaner Z, Inostroza J, Costa-Carvalho BT, de Albuquerque JAT, Garcia-Ortiz H, Orozco L, Ozcelik T, Abid A, Rhorfi IA, Souhi H, Amrani HN, Zegmout A, Geissmann F, Michnick SW, Muller-Fleckenstein I, Fleckenstein B, Puel A, Ciancanelli MJ, Marr N, Abolhassani H, Balcells ME, Condino-Neto A, Strickler A, Abarca K, Teuscher C, Ochs HD, Reisli I, Sayar EH, El-Baghdadi J, Bustamante J, Hammarström L, Tangye SG, Pellegrini S, Quintana-Murci L, Abel L, Casanova JL (2018) Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci Immunol 3:eaau8714. 10.1126/sciimmunol.aau8714 - PMC - PubMed
-
- Gorman JA, Hundhausen C, Kinsman M, Arkatkar T, Allenspach EJ, Clough C, West SE, Thomas K, Eken A, Khim S, Hale M, Oukka M, Jackson SW, Cerosaletti K, Buckner JH, Rawlings DJ (2019) The TYK2-P1104A autoimmune protective variant limits coordinate signals required to generate specialized T cell subsets. Front Immunol 10:44. 10.3389/fimmu.2019.00044 - PMC - PubMed
-
- Kreins AY, Ciancanelli MJ, Okada S, Kong XF, Ramírez-Alejo N, Kilic SS, El Baghdadi J, Nonoyama S, Mahdaviani SA, Ailal F, Bousfiha A, Mansouri D, Nievas E, Ma CS, Rao G, Bernasconi A, Sun Kuehn H, Niemela J, Stoddard J, Deveau P, Cobat A, El Azbaoui S, Sabri A, Lim CK, Sundin M, Avery DT, Halwani R, Grant AV, Boisson B, Bogunovic D, Itan Y, Moncada-Velez M, Martinez-Barricarte R, Migaud M, Deswarte C, Alsina L, Kotlarz D, Klein C, Muller-Fleckenstein I, Fleckenstein B, Cormier-Daire V, Rose-John S, Picard C, Hammarstrom L, Puel A, Al-Muhsen S, Abel L, Chaussabel D, Rosenzweig SD, Minegishi Y, Tangye SG, Bustamante J, Casanova JL, Boisson-Dupuis S (2015) Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J Exp Med 212:1641–1662. 10.1084/jem.20140280 - PMC - PubMed
-
- Li Z, Gakovic M, Ragimbeau J, Eloranta ML, Rönnblom L, Michel F, Pellegrini S (2013) Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol 190:2335–2344. 10.4049/jimmunol.1203118 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

