NGS detection of gene rearrangements and METexon14 mutations in liquid biopsy of advanced NSCLC patients: A study of two Italian centers
- PMID: 40027148
- PMCID: PMC11863814
- DOI: 10.1016/j.jlb.2024.100143
NGS detection of gene rearrangements and METexon14 mutations in liquid biopsy of advanced NSCLC patients: A study of two Italian centers
Abstract
Introduction: ctDNA is a useful tool for NGS molecular profiling in advanced NSCLC patients. Its clinical applicability in patients with gene rearrangements is still limited due to a lower detection rate of these types of alterations compared to single SNVs or small indels. To this purpose, we performed a study in two Italian centers to assess the concordance between tissue and plasma samples in the detection of genes fusions (ALK, ROS, RET) and METexon14 mutations in advanced NSCLC patients.
Methods: Patients with a histological diagnosis of oncogene addicted (ALK, ROS1, RET positive or METexon14 mutated) advanced NSCLC were enrolled at the time of first line of TKI treatment. Plasma samples were harvested before the start of TKI treatment and NGS analysis on ctDNA samples using the AVENIO ctDNA Expanded kit was performed. The Positive Percent Agreement (PPA) between tissue and plasma was calculated.
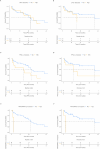
Results: Fifty-eight rearranged or METexon14 mutated NSCLC patients were included and 57 ctDNA samples were successfully sequenced. An overall PPA of 37% (21/57) was obtained, with a best performance for RET fusion (80%), intermediate for METexon14 skipping mutations (40%) and ALK rearranged (36%) and a worst one for ROS1 rearranged samples (18%). We found TP53, APC and SMAD4 as most prevalent co-mutated genes (21%, 12% and 10% of patients, respectively). Among different factors considered, increased driver detection rate in patients with extra-thoracic metastases (p = 0.0049) was observed. Significantly shorter survival was observed in patients harboring co-occurring KRAS/NRAS mutations in ctDNA.
Conclusions: ctDNA testing to detect oncogenic fusions or METexon14 mutations in advanced NSCLC patients is useful, even if type of gene alterations and clinical characteristics could influence the driver detection rate. Liquid biopsy represents a complementary tool to tissue genotyping, however more sensitive approaches for gene fusions and METexon14 detection are needed to implement its strength and reliability.
Keywords: Advanced NSCLC; Concordance rate; Gene rearrangements; Liquid biopsy; METexon14 mutations; ctDNA.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Giulia Pasello reports financial support and article publishing charges were provided by Veneto Oncology Institute. Marcello Tiseo reports financial support was provided by University Hospital of Parma. Marcello Tiseo reports a relationship with Astra-Zeneca, Pfizer, Eli-Lilly, BMS, Novartis, Roche, MSD, Boehringer Ingelheim, Otsuka, Takeda, Pierre Fabre, Amgen, Merck, Sanofi, Janssen, Daiichi Sankyo that includes: consulting or advisory and speaking and lecture fees. Marcello Tiseo reports a relationship with Astra-Zeneca, Boehringer Ingelheim. That includes: funding grants. Giulia Pasello reports a relationship with Astra-Zeneca, Amgen, BMS, Eli Lilly, Janssen, MSD, Novartis, Roche that includes: consulting or advisory and speaking and lecture fees. Giulia Pasello reports a relationship with Astra-Zeneca, Roche, MSD that includes: funding grants. Laura Bonanno reports a relationship with Astra-Zeneca, MSD, BMS, Roche, Novartis, Eli-Lilly that includes: consulting or advisory and speaking and lecture fees. Laura Bonanno reports a relationship with Astra-Zeneca. That includes: funding grants. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

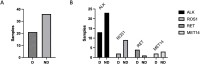


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

