Novel Insights into PGM2L1 as a Prognostic Biomarker in Cholangiocarcinoma: Implications for Metabolic Reprogramming and Tumor Microenvironment Modulation
- PMID: 40027181
- PMCID: PMC11866533
- DOI: 10.7150/ijms.106566
Novel Insights into PGM2L1 as a Prognostic Biomarker in Cholangiocarcinoma: Implications for Metabolic Reprogramming and Tumor Microenvironment Modulation
Abstract
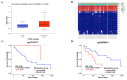
Cholangiocarcinoma (CCA) is a highly lethal malignancy and the most common adenocarcinoma of the hepatobiliary system. PGM2L1 belongs to the α-D-phosphohexomutase superfamily and functions as a glucose 1,6-bisphosphate (G16BP) synthase. There is growing evidence to provide an association its function to cancer metabolism and progression. However, the molecular mechanisms of PGM2L1 in CCA development remain lacking in evidence. In this study, we found that CCA patients with high PGM2L1 expression had the poorest prognosis. We identified two methylation sites (cg15214137 and cg03699633) within the PGM2L1 gene and their prognostic relevance. We further investigated the relationship between PGM2L1 expression and tumor-infiltrating immune cells, with a particular focus on neutrophils in CCA. Functional enrichment analyses further revealed that high PGM2L1 expression was associated with the Wnt signaling pathway, glycolytic metabolism, and the recruitment of neutrophils. Collectively, these findings suggest that PGM2L1 may serve as an independent prognostic biomarker and is closely linked to tumor immune infiltration and metabolic reprogramming in CCA.
Keywords: PGM2L1; cholangiocarcinoma; glycolytic metabolism; tumor-infiltrating immune cells.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

