This is a preprint.
Coordinated macrophage and T cell interactions mediate response to checkpoint blockade in colorectal cancer
- PMID: 40027748
- PMCID: PMC11870396
- DOI: 10.1101/2025.02.12.637954
Coordinated macrophage and T cell interactions mediate response to checkpoint blockade in colorectal cancer
Abstract
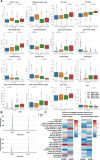
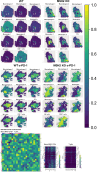
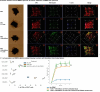
Mismatch repair deficiency (MMRd), either due to inherited or somatic mutation, is prevalent in colorectal cancer (CRC) and other cancers. While anti-PD-1 therapy is utilized in both local and advanced disease, up to 50% of MMRd CRC fail to respond. Using animal and human models of MMRd, we determined that interactions between MHC+ C1Q+ CXCL9+ macrophages and TCF+ BHLHE40+ PRF1+ T cell subsets are associated with control of MMRd tumor growth, during anti-PD-1 treatment. In contrast, resistance is associated with upregulation of TIM3, LAG3, TIGIT, and PD-1 expression on T cells, and infiltration of the tumor with immunosuppressive TREM2+ macrophages and monocytes. By combining anti-PD-1 with anti-LAG3/CTLA4/TREM2, up to 100% tumor eradication was achieved in MMRd CRC and remarkably, in >70% in MMRp CRC. This study identifies key T cell and macrophage subsets mediating the efficacy of immunotherapy in overcoming immune escape in both MMRd and MMRp CRC settings.
Keywords: CTLA-4; LAG3; Mismatch repair deficiency; PD-1; TREM2; immune resistance.
Conflict of interest statement
Declaration of Interests M.B. is a Parker Scholar with the Parker Institute for Cancer Immunotherapy. R.S. is a co-inventors on a patent (US11230599/EP4226944A3) filed by MSKCC for using TMB to predict immunotherapy response, licensed to Personal Genome Diagnostics (PGDx). N.B. is an extramural member of the Parker Institute for Cancer Immunotherapy. N.B. received research support from Harbour Biomed Sciences, stock option from BreakBio, serves as Advisor/Board Member at Curevac, serves as Advisor/Board Member and received stock option from Genotwin and DC Prime, serves as Advisor/Board Member and received equity from Cell BioEngines, received hold stocks from Barinthus, serves as consultant and grant recipient at Merck Research Laboratories, received a drug product from Oncovir and serves at scientific advisory board and received stock options from Aikium. The other authors have not declared any competing interests.
Figures




















References
-
- Gryfe R. et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 342, 69–77 (2000). - PubMed
-
- Yamamoto H. et al. Gastric cancers of the microsatellite mutator phenotype display characteristic genetic and clinical features. Gastroenterology 116, 1348–1357 (1999). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
