Overlap of Formalin-Fixed Paraffin-Embedded and Fresh-Frozen Matched Tissues for Proteomics and Phosphoproteomics
- PMID: 40028131
- PMCID: PMC11865994
- DOI: 10.1021/acsomega.4c09289
Overlap of Formalin-Fixed Paraffin-Embedded and Fresh-Frozen Matched Tissues for Proteomics and Phosphoproteomics
Abstract
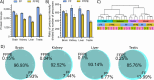
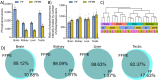
Many liquid chromatography-mass spectrometry (LC-MS) studies have compared formalin-fixed paraffin-embedded (FFPE) tissues with matched fresh-frozen (FF) tissues to examine the effect of preservation techniques on the proteome; however, few studies have included the phosphoproteome. A high degree of overlap and correlation between the two preservation techniques would demonstrate the importance of FFPE tissues as a valuable biomedical resource. Our aim was to quantitatively compare the proteome and phosphoproteome of matched FFPE and FF tissues using data-independent acquisition LC-MS. Four organs from three rats were cut in half to produce matched FFPE and FF tissue pairs. Excellent overlaps of 85-97% for the proteome and 82-98% for the phosphoproteome were observed, depending on the organ type, between the two preservation techniques. Most of the unique identifications were found in FF with less than 0.3% being unique to FFPE tissues. Strong agreement between FFPE and FF matched tissue pairs was observed with Pearson correlation coefficients of 0.93-0.97 and 0.79-0.87 for the proteome and phosphoproteome, respectively. Digestion efficiency was slightly higher in FFPE (92-94%) than in FF tissues (86-89%), and a search of a data subset for formaldehyde induced chemical modifications revealed that only 0.05% of precursors were unique to FFPE tissues. This suggests that with quality sample preparation methods it is not necessary to include formaldehyde induced chemical modifications when analyzing FFPE tissues. We attribute the lower number of identifications in FFPE tissues to inaccurate peptide quantitation, which resulted in a lower MS peptide load and tryptic peptide enrichment load. Our results demonstrate that both proteomic and phosphoproteomic analyses of FFPE and FF tissues are highly comparable and highlight the suitability of FFPE tissues for both proteomic and phosphoproteomic analysis.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues.J Proteomics. 2021 Mar 20;235:104117. doi: 10.1016/j.jprot.2021.104117. Epub 2021 Jan 14. J Proteomics. 2021. PMID: 33453434 Free PMC article. Review.
-
Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis.Mol Cell Proteomics. 2009 Aug;8(8):1988-98. doi: 10.1074/mcp.M800518-MCP200. Epub 2009 May 24. Mol Cell Proteomics. 2009. PMID: 19467989 Free PMC article.
-
Proteomics Analysis of Formalin Fixed Paraffin Embedded Tissues in the Investigation of Prostate Cancer.J Proteome Res. 2020 Jul 2;19(7):2631-2642. doi: 10.1021/acs.jproteome.9b00587. Epub 2019 Nov 18. J Proteome Res. 2020. PMID: 31682457
-
Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue.PLoS One. 2011 Feb 11;6(2):e17163. doi: 10.1371/journal.pone.0017163. PLoS One. 2011. PMID: 21347257 Free PMC article.
-
LC-MS bioanalysis of protein biomarkers and protein therapeutics in formalin-fixed paraffin-embedded tissue specimens.Bioanalysis. 2024 Feb;16(4):245-258. doi: 10.4155/bio-2023-0210. Epub 2024 Jan 16. Bioanalysis. 2024. PMID: 38226835 Review.
Cited by
-
Integrative Spatial Proteomics and Single-Cell RNA Sequencing Unveil Molecular Complexity in Rheumatoid Arthritis for Novel Therapeutic Targeting.Proteomes. 2025 May 22;13(2):17. doi: 10.3390/proteomes13020017. Proteomes. 2025. PMID: 40559990 Free PMC article.
References
-
- Ostasiewicz P.; Zielinska D. F.; Mann M.; Wisniewski J. R. Proteome, Phosphoproteome, and N-Glycoproteome Are Quantitatively Preserved in Formalin-Fixed Paraffin-Embedded Tissue and Analyzable by High-Resolution Mass Spectrometry. J. Proteome. Res. 2010, 9 (7), 3688–3700. 10.1021/pr100234w. - DOI - PubMed
-
- Gamez-Pozo A.; Sanchez-Navarro I.; Calvo E.; Diaz E.; Miguel-Martin M.; Lopez R.; Agullo T.; Camafeita E.; Espinosa E.; Lopez J. A.; et al. Protein phosphorylation analysis in archival clinical cancer samples by shotgun and targeted proteomics approaches. Mol. Biosyst 2011, 7 (8), 2368–2374. 10.1039/c1mb05113j. - DOI - PubMed
-
- Gundisch S.; Hauck S.; Sarioglu H.; Schott C.; Viertler C.; Kap M.; Schuster T.; Reischauer B.; Rosenberg R.; Verhoef C.; et al. Variability of protein and phosphoprotein levels in clinical tissue specimens during the preanalytical phase. J. Proteome. Res. 2012, 11 (12), 5748–5762. 10.1021/pr300560y. - DOI - PubMed
-
- Vitorio J. G.; Duarte-Andrade F. F.; Pereira T.; Melo-Braga M. N.; Canuto G. A. B.; Macedo A. N.; Lebron Y. A. R.; Moreira V. R.; Felicori L. F.; Lange L. C.; et al. Integrated proteomics, phosphoproteomics and metabolomics analyses reveal similarities among giant cell granulomas of the jaws with different genetic mutations. J. Oral. Pathol. Med. 2022, 51 (7), 666–673. 10.1111/jop.13327. - DOI - PubMed
-
From NLM Medline.
-
- Duarte-Andrade F. F.; Pereira T.; Vitorio J. G.; Diniz M. G.; Amorim L. S. D.; Nawrocki A.; Felicori L. F.; De Marco L.; Gomes C. C.; Larsen M. R.; et al. Quantitative proteomic study reveals differential expression of matricellular proteins between fibrous dysplasia and cemento-ossifying fibroma pathogenesis. J. Oral. Pathol. Med. 2022, 51 (4), 405–412. 10.1111/jop.13282. - DOI - PubMed
-
From NLM Medline.
LinkOut - more resources
Full Text Sources
