Optimal input DNA thresholds for genome skimming in marine crustacean zooplankton
- PMID: 40028195
- PMCID: PMC11871894
- DOI: 10.7717/peerj.19054
Optimal input DNA thresholds for genome skimming in marine crustacean zooplankton
Abstract
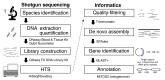
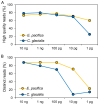
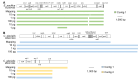
Crustacean zooplanktons are key secondary and tertiary producers in marine ecosystems, yet their genomic resources remain poorly understood. To advance biodiversity research on crustacean zooplankton, this study evaluated the effectiveness of genome skimming, a method that assembles genetic regions, including mitogenome, from shotgun genome sequencing data. Because the small amount of DNA available is a limitation in zooplankton genetics, different input DNA amounts (1 pg-10 ng) were prepared for library construction for genome skimming using two large species: Euphausia pacifica (Euphausiacea) and Calanus glacialis (Copepoda). Additionally, de novo assembly was used to obtain long contigs from short reads because reference-guided assembly can not be applied to all crustacean zooplankton. Evaluation of the raw sequence reads showed increased proportions of high-quality and distinct reads (low duplication levels) for large DNA inputs. By contrast, low sequence quality and high sequence duplication were observed for ≤ 10 pg DNA samples, owing to increased DNA amplification cycles. Complete mitogenomes, including all 37 genes, were successfully retrieved for ≥ 10 pg (E. pacifica) and ≥ 100 pg (C. glacialis) of DNA. Despite the large estimated genome sizes of these zooplankton species, only ≥ 1 and ≥ 3 M reads were sufficient for mitogenome assembly for E. pacifica and C. glacialis, respectively. Nuclear ribosomal repeats and histone 3 were identified in the assembled contigs. As obtaining sufficient DNA amounts (≥ 100 pg) is feasible even from small crustacean zooplankton, genome skimming is a powerful approach for robust phylogenetics and population genetics in marine zooplankton.
Keywords: Copepoda; Euphausiacea; Genome skimming; Mitogenome; Zooplankton.
©2025 Hirai.
Conflict of interest statement
The author declares there are no competing interests.
Figures

Similar articles
-
Mitochondrial genomes of the key zooplankton copepods Arctic Calanus glacialis and North Atlantic Calanus finmarchicus with the longest crustacean non-coding regions.Sci Rep. 2017 Oct 20;7(1):13702. doi: 10.1038/s41598-017-13807-0. Sci Rep. 2017. PMID: 29057900 Free PMC article.
-
The complete mitochondrial genome sequence of Euphausia pacifica (Malacostraca: Euphausiacea) reveals a novel gene order and unusual tandem repeats.Genome. 2011 Nov;54(11):911-22. doi: 10.1139/g11-053. Epub 2011 Oct 21. Genome. 2011. PMID: 22017501
-
Skimming for barcodes: rapid production of mitochondrial genome and nuclear ribosomal repeat reference markers through shallow shotgun sequencing.PeerJ. 2022 Aug 5;10:e13790. doi: 10.7717/peerj.13790. eCollection 2022. PeerJ. 2022. PMID: 35959477 Free PMC article.
-
Mitochondrial metagenomics: letting the genes out of the bottle.Gigascience. 2016 Mar 22;5:15. doi: 10.1186/s13742-016-0120-y. eCollection 2016. Gigascience. 2016. PMID: 27006764 Free PMC article. Review.
-
[Mitogenome assembly strategies and software applications in the genome era].Yi Chuan. 2019 Nov 20;41(11):979-993. doi: 10.16288/j.yczz.19-227. Yi Chuan. 2019. PMID: 31735702 Review. Chinese.
References
-
- Baker ADC, Boden BP, Brinton E. Practical guide to the euphausiids of the world. London: British Museum (Natural History); 1990.
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

