Extracellular vesicles of Limosilactobacillus fermentum SLAM216 ameliorate skin symptoms of atopic dermatitis by regulating gut microbiome on serotonin metabolism
- PMID: 40028723
- PMCID: PMC11881872
- DOI: 10.1080/19490976.2025.2474256
Extracellular vesicles of Limosilactobacillus fermentum SLAM216 ameliorate skin symptoms of atopic dermatitis by regulating gut microbiome on serotonin metabolism
Abstract
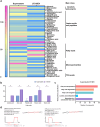
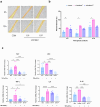
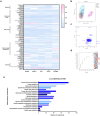
Atopic dermatitis (AD) is a globally prevalent chronic inflammatory skin disorder. Its pathogenesis remains incompletely understood, resulting in considerable therapeutic challenges. Recent studies have highlighted the significance of the interaction between AD and gut microbiome. In this study, we investigated the effects of probiotic-derived extracellular vesicles on AD. Initially, we isolated and characterized extracellular vesicles from Limosilactobacillus fermentum SLAM 216 (LF216EV) and characterized their composition through multi-omics analysis. Gene ontology (GO) and pathway analysis classified LF216EV proteins into biological processes, molecular functions, and cellular components. Importantly, specific abundance in linoleic, oleic, palmitic, sebacic, and stearic acids indicating upregulated fatty acid metabolism were observed by metabolomic analysis. Furthermore, featured lipid profiling including AcylGlcADG and ceramide were observed in LF216EV. Importantly, in an atopic dermatitis-like cell model induced by TNFα/IFNγ, LF216EV significantly modulated the expression of immune regulatory genes (TSLP, TNFα, IL-6, IL-1β, and MDC), indicating its potential functionality in atopic dermatitis. LF216EV alleviated AD-like phenotypes, such as redness, scaling/dryness, and excoriation, induced by DNCB. Histopathological analysis revealed that LF216EV decreased epidermal thickness and mast cell infiltration in the dermis. Furthermore, LF216EV administration reduced mouse scratching and depression-related behaviors, with a faster onset than the classical treatment with dexamethasone. In the quantitative real-time polymerase chain reaction (qRT-PCR) analysis, we observed a significant increase in the expression levels of htrb2c, sert, and tph-1, genes associated with serotonin, in the skin and gut of the LF216EV-treated group, along with a significant increase in the total serum serotonin levels. Gut microbiome analysis of the LF216EV-treated group revealed an altered gut microbiota profile. Correlation analysis revealed that the genera Limosilactobacillus and Desulfovibrio were associated with differences in the intestinal metabolites, including serotonin. Our findings demonstrate that LF216EV mitigates AD-like symptoms by promoting serotonin synthesis through the modulation of gut microbiota and metabolome composition.
Keywords: Atopic dermatitis; extracellular vesicle; gut microbiome; gut-skin axis; multi-omics analysis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures








References
-
- Reddel S, Del Chierico F, Quagliariello A, Giancristoforo S, Vernocchi P, Russo A, Fiocchi A, Rossi P, Putignani L, El Hachem M. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep. 2019;9(1):4996. doi:10.1038/s41598-019-41149-6. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
