A conformational fingerprint for amyloidogenic light chains
- PMID: 40028903
- PMCID: PMC11875538
- DOI: 10.7554/eLife.102002
A conformational fingerprint for amyloidogenic light chains
Abstract
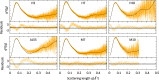
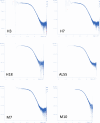
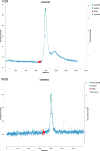
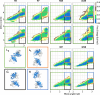
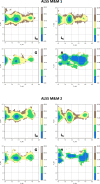
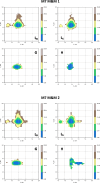
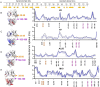
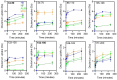
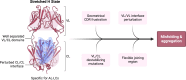
Both immunoglobulin light-chain (LC) amyloidosis (AL) and multiple myeloma (MM) share the overproduction of a clonal LC. However, while LCs in MM remain soluble in circulation, AL LCs misfold into toxic-soluble species and amyloid fibrils that accumulate in organs, leading to distinct clinical manifestations. The significant sequence variability of LCs has hindered the understanding of the mechanisms driving LC aggregation. Nevertheless, emerging biochemical properties, including dimer stability, conformational dynamics, and proteolysis susceptibility, distinguish AL LCs from those in MM under native conditions. This study aimed to identify a2 conformational fingerprint distinguishing AL from MM LCs. Using small-angle X-ray scattering (SAXS) under native conditions, we analyzed four AL and two MM LCs. We observed that AL LCs exhibited a slightly larger radius of gyration and greater deviations from X-ray crystallography-determined or predicted structures, reflecting enhanced conformational dynamics. SAXS data, integrated with molecular dynamics simulations, revealed a conformational ensemble where LCs adopt multiple states, with variable and constant domains either bent or straight. AL LCs displayed a distinct, low-populated, straight conformation (termed H state), which maximized solvent accessibility at the interface between constant and variable domains. Hydrogen-deuterium exchange mass spectrometry experimentally validated this H state. These findings reconcile diverse experimental observations and provide a precise structural target for future drug design efforts.
Keywords: amyloidogenic light chain; biochemistry; chemical biology; conformational dynamics; hydrogen deuterium exchange; molecular biophysics; molecular dynamics; none; small-angle X-ray scattering; structural biology.
© 2024, Paissoni, Puri et al.
Conflict of interest statement
CP, SP, LB, MS, MM, RR, VS, FB, MN, GM, GP, SH, SR, CC No competing interests declared
Figures


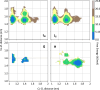
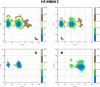
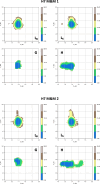
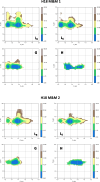












Update of
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
-
- Achour A, Broggini L, Han X, Sun R, Santambrogio C, Buratto J, Visentin C, Barbiroli A, De Luca CMG, Sormanni P, Moda F, De Simone A, Sandalova T, Grandori R, Camilloni C, Ricagno S. Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity. The FEBS Journal. 2020;287:546–560. doi: 10.1111/febs.15046. - DOI - PubMed
-
- Ahmed MC, Skaanning LK, Jussupow A, Newcombe EA, Kragelund BB, Camilloni C, Langkilde AE, Lindorff-Larsen K. Refinement of α-synuclein ensembles against SAXS data: comparison of force fields and methods. Frontiers in Molecular Biosciences. 2021;8:654333. doi: 10.3389/fmolb.2021.654333. - DOI - PMC - PubMed

