Stabilization of the catalytically active structure of a molybdenum-dependent formate dehydrogenase depends on a highly conserved lysine residue
- PMID: 40028997
- PMCID: PMC12176258
- DOI: 10.1111/febs.70048
Stabilization of the catalytically active structure of a molybdenum-dependent formate dehydrogenase depends on a highly conserved lysine residue
Abstract
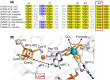
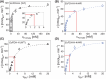
Molybdenum-dependent formate dehydrogenases (Mo-FDHs) reversibly catalyze the interconversion of CO2 and formate, and therefore may be utilized for the development of innovative energy storage and CO2 utilization concepts. Mo-FDHs contain a highly conserved lysine residue in the vicinity of a catalytically active molybdenum (Mo) cofactor and an electron-transferring [4Fe-4S] cluster. In order to elucidate the function of the conserved lysine, we substituted the residue Lys44 of Escherichia coli formate dehydrogenase H (EcFDH-H) with structurally and chemically diverse amino acids. Enzyme kinetic analysis of the purified EcFDH-H variants revealed the Lys-to-Arg substitution as the only amino acid exchange that retained formate oxidation catalytic activity, amounting to 7.1% of the wild-type level. Ultraviolet-visible (UV-Vis) spectroscopic analysis indicated that >90% of the [4Fe-4S] cluster was lost in the case of EcFDH-H variants -K44E and -K44M, whereas the cluster occupancy of the K44R variant decreased by merely 4.5%. Furthermore, the K44R substitution resulted in a slight decrease in its melting temperature and a significant formate affinity decrease, apparent as a 32-fold Km value increase. Consistent with these findings, molecular dynamics simulations predicted an increase in the backbone and cofactor mobility as a result of the K44R substitution. These results are consistent with the conserved lysine being essential for stabilizing the catalytically active structures in EcFDH-H and may support engineering efforts on Mo-FDHs to design more efficient biocatalysts for CO2 reduction.
Keywords: CO2 utilization; lysine; molybdenum‐dependent formate dehydrogenase; redox cofactor; site‐directed mutagenesis.
© 2025 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Schwarz G, Mendel RR & Ribbe MW (2009) Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847. - PubMed
-
- Leimkühler S (2020) The biosynthesis of the molybdenum cofactors in Escherichia coli . Environ Microbiol 22, 2007–2026. - PubMed
-
- Mendel RR & Leimkühler S (2015) The biosynthesis of the molybdenum cofactors. J Biol Inorg Chem 20, 337–347. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

