De Novo Serine Synthesis Is a Metabolic Vulnerability That Can Be Exploited to Overcome Sunitinib Resistance in Advanced Renal Cell Carcinoma
- PMID: 40029310
- PMCID: PMC12079101
- DOI: 10.1158/0008-5472.CAN-24-1393
De Novo Serine Synthesis Is a Metabolic Vulnerability That Can Be Exploited to Overcome Sunitinib Resistance in Advanced Renal Cell Carcinoma
Abstract
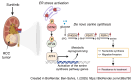
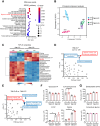
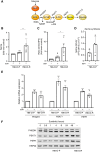
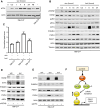
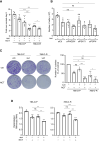
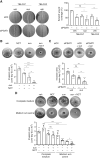
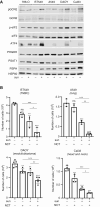
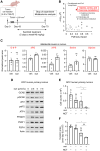
Sunitinib is an oral tyrosine kinase inhibitor used in treating advanced renal cell carcinoma (RCC) that exhibits significant efficacy but faces resistance in 30% of patients. Identifying the molecular mechanisms underlying resistance could enable the development of strategies to enhance sunitinib sensitivity. In this study, we showed that sunitinib induces a metabolic shift leading to increased serine synthesis in RCC cells. Activation of the GCN2-ATF4 stress response pathway was identified as the mechanistic link between sunitinib treatment and elevated serine production. The increased serine biosynthesis supported nucleotide synthesis and sustained cell proliferation, migration, and invasion following sunitinib treatment. Inhibiting key enzymes in the serine synthesis pathway, such as phosphoglycerate dehydrogenase and phosphoserine aminotransferase 1, enhanced the sensitivity of resistant cells to sunitinib. Beyond RCC, similar activation of serine synthesis following sunitinib treatment occurred in a variety of other cancer types, suggesting a shared adaptive response to sunitinib therapy. Together, this study identifies the de novo serine synthesis pathway as a potential target to overcome sunitinib resistance, offering insights into therapeutic strategies applicable across diverse cancer contexts. Significance: Sunitinib treatment induces metabolic reprogramming to provide essential metabolite building blocks for tumor survival, resistance, and progression by upregulating serine biosynthesis, which represents a targetable dependency to enhance therapeutic efficacy.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures
References
-
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999;399:271–5. - PubMed
-
- Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2002;2:673–82. - PubMed
MeSH terms
Substances
Grants and funding
- Emergence/Canceropôle PACA (Canceropole PACA)
- Fondation de France (Foundation of France)
- DBG-23-1039959-01-TBE/American Cancer Society (ACS)
- R01 GM143334/GM/NIGMS NIH HHS/United States
- R01GM135587/Foundation for the National Institutes of Health (FNIH)
- 2019/Canceropôle PACA (Canceropole PACA)
- Fondation pour la Recherche Médicale (FRM)
- R01GM143334-04/Foundation for the National Institutes of Health (FNIH)
- R01 GM135587/GM/NIGMS NIH HHS/United States
- ARCAGEING202302000633/Fondation ARC pour la Recherche sur le Cancer (ARC)
- ANR-21-CE14-0008-0/Agence Nationale de la Recherche (ANR)
- Ligue Contre le Cancer (French League Against Cancer)
- Credits incitatifs/UNICA
- PJA20191209349/Fondation ARC pour la Recherche sur le Cancer (ARC)
- French ministry of research
LinkOut - more resources
Full Text Sources
Medical
Research Materials

