CRISPR-mediated detection of Pneumocystis transcripts in bronchoalveolar, oropharyngeal, and serum specimens for Pneumocystis pneumonia diagnosis
- PMID: 40029713
- PMCID: PMC11996908
- DOI: 10.1172/JCI177241
CRISPR-mediated detection of Pneumocystis transcripts in bronchoalveolar, oropharyngeal, and serum specimens for Pneumocystis pneumonia diagnosis
Abstract
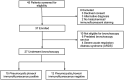
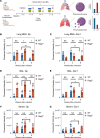
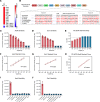
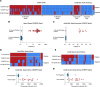
BACKGROUNDPneumocystis jirovecii pneumonia (PCP) is a leading cause of fungal pneumonia, but its diagnosis primarily relies on invasive bronchoalveolar lavage (BAL) specimens that are difficult to obtain. Oropharyngeal swabs and serum could improve the PCP diagnostic workflow, and we hypothesized that CRISPR could enhance assay sensitivity to allow robust P. jirovecii diagnosis using swabs and serum. Herein, we describe the development of an ultrasensitive RT-PCR-coupled CRISPR assay with high active-infection specificity in infant swabs and adult BAL and serum.METHODSMouse analyses employed an RT-PCR CRISPR assay to analyze P. murina transcripts in WT and Rag2-/- mouse lung RNA, BAL, and serum at 2-, 4-, and 6-weeks after infection. Human studies used an optimized RT-PCR CRISPR assay to detect P. jirovecii transcripts in infant oropharyngeal swab samples, adult serum, and adult BAL specimens from patients who were infected with P. jirovecii and those who were not.RESULTSThe P. murina assays sensitively detected Pneumocystis RNA in the serum of infected mice throughout infection. Oropharyngeal swab CRISPR assay results identified infants infected with P. jirovecii with greater sensitivity (96.3% versus 66.7%) and specificity (100% versus 90.6%) than RT-qPCR compared with mitochondrial large subunit rRNA gene (mtLSU) standard marker, and CRISPR results achieved higher sensitivity than RT-qPCR results (93.3% versus 26.7%) in adult serum specimens.CONCLUSIONSince swabs are routinely collected in pediatric patients with pneumonia and serum is easier to obtain than BAL, this assay approach could improve the accuracy and timing of pediatric and adult Pneumocystis diagnosis by achieving specificity for active infection and potentially avoiding the requirement for BAL specimens.FUNDINGThe work was supported by the NIH (R01AI120033), NHLBI (R35HL139930), the Louisiana Board of Regents Endowed Chairs for Eminent Scholars program, and by research funding provided by National Institute of Allergy and Infectious Diseases (NIAID) (R01AI144168, R01AI175618, R01AI173021). This research was also funded by the NIHR (project 134342) using UK aid from the UK government to support global health research.
Keywords: Diagnostics; Infectious disease; Mitochondria; Mouse models; Pulmonology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

