Identification of 2 novel noncoding variants in patients with Diamond-Blackfan anemia syndrome by whole genome sequencing
- PMID: 40029997
- PMCID: PMC12144513
- DOI: 10.1182/bloodadvances.2024015347
Identification of 2 novel noncoding variants in patients with Diamond-Blackfan anemia syndrome by whole genome sequencing
Abstract
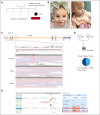
Diamond-Blackfan anemia syndrome (DBAS) is a rare congenital disorder with variable penetrance and expressivity and is characterized by pure red cell aplasia that typically manifests as early-onset chronic macrocytic or normocytic anemia and is often associated with other congenital anomalies. DBAS is etiologically heterogeneous with >20 known DBAS-associated genes that encode small and large ribosomal protein subunits and an inheritance pattern that is largely autosomal dominant or sporadic. We report 2 DBAS cases with previous negative genetic testing, which included targeted gene panels, karyotype analysis, chromosome breakage analysis, and whole exome sequencing. Although clinical whole genome sequencing (WGS) was initially negative, in-depth reanalysis identified 2 novel noncoding variants in the RPS gene family, namely a maternally inherited splicing variant at the end of the first noncoding exon in RPS7 (NM_001011.4, c.-19G>C) in family 1 and a deep intronic de novo variant in RPS19 (NM_001022.4, c.172+350C>T) in family 2. In family 1, several maternal relatives were identified who shared the same variant through cascade testing; clinically, they exhibited variable degrees of anemia and elevated erythrocyte adenosine deaminase activity, a marker for DBAS. RNA sequencing analysis demonstrated deleterious functional consequences for both noncoding variants. In case 1, hematopoietic stem cell transplant with an unaffected matched sibling donor who did not carry the variant successfully cured the congenital anemia. This study identified novel noncoding variants and underscores the clinical utility of WGS in accelerating diagnosis and improving care for rare genetic disorders, particularly when timely treatment decisions are critically important.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.W. is Pathology and Laboratory Medicine, Henry Ford Hospital, Detroit, MI.
Figures



References
-
- Sieff C. In: GeneReviews. Adam MP, Feldman J, Mirzaa GM, et al., editors. University of Washington; 1993. Diamond-Blackfan anemia.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

