Recessive genetic contribution to congenital heart disease in 5,424 probands
- PMID: 40030011
- PMCID: PMC11912448
- DOI: 10.1073/pnas.2419992122
Recessive genetic contribution to congenital heart disease in 5,424 probands
Abstract
Variants with large effect contribute to congenital heart disease (CHD). To date, recessive genotypes (RGs) have commonly been implicated through anecdotal ascertainment of consanguineous families and candidate gene-based analysis; the recessive contribution to the broad range of CHD phenotypes has been limited. We analyzed whole exome sequences of 5,424 CHD probands. Rare damaging RGs were estimated to contribute to at least 2.2% of CHD, with greater enrichment among laterality phenotypes (5.4%) versus other subsets (1.4%). Among 108 curated human recessive CHD genes, there were 66 RGs, with 54 in 11 genes with >1 RG, 12 genes with 1 RG, and 85 genes with zero. RGs were more prevalent among offspring of consanguineous union (4.7%, 32/675) than among nonconsanguineous probands (0.7%, 34/4749). Founder variants in GDF1 and PLD1 accounted for 74% of the contribution of RGs among 410 Ashkenazi Jewish probands. We identified genome-wide significant enrichment of RGs in C1orf127, encoding a likely secreted protein expressed in embryonic mouse notochord and associated with laterality defects. Single-cell transcriptomes from gastrulation-stage mouse embryos revealed enrichment of RGs in genes highly expressed in the cardiomyocyte lineage, including contractility-related genes MYH6, UNC45B, MYO18B, and MYBPC3 in probands with left-sided CHD, consistent with abnormal contractile function contributing to these malformations. Genes with significant RG burden account for 1.3% of probands, more than half the inferred total. These results reveal the recessive contribution to CHD, and indicate that many genes remain to be discovered, with each likely accounting for a very small fraction of the total.
Keywords: congenital heart disease; exome-sequencing; genomics; human genetics.
Conflict of interest statement
Competing interests statement:M.Y. is a cofounder and consultant for Fabric Genomics Inc., M.Y. has stock in Fabric Genomics.
Figures
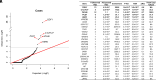

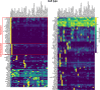

References
MeSH terms
Substances
Grants and funding
- U01 HL128711/HL/NHLBI NIH HHS/United States
- RM1HG011014/HHS | NIH | National Human Genome Research Institute (NHGRI)
- Howard Hughes Medical Institute/Howard Hughes Medical Institute (HHMI)
- U01 HL098162/HL/NHLBI NIH HHS/United States
- UL1 TR000003/TR/NCATS NIH HHS/United States
- 1UG1HL135680-01/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- CDI-FR-2021-926/Children's Discovery Institute (CDI)
- UO1-HL128711/HHS | NIH | NHLBI | Division of Intramural Research (DIR)
- UG1 HL135680/HL/NHLBI NIH HHS/United States
- UO1-HL098147/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R03 HD100883/HD/NICHD NIH HHS/United States
- RM1 HG011014/HG/NHGRI NIH HHS/United States
- U01 HL098153/HL/NHLBI NIH HHS/United States
- U01 HL131003/HL/NHLBI NIH HHS/United States
- 5U54HG006504/HHS | NIH | National Human Genome Research Institute (NHGRI)
- UO1-HL098162/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- U01 HL153009/HL/NHLBI NIH HHS/United States
- R00 HL143036/HL/NHLBI NIH HHS/United States
- HL157653/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- HL157653/HHS | NIH | NHLBI | Division of Intramural Research (DIR)
- U54 HG006504/HG/NHGRI NIH HHS/United States
- R00HL143036-02/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 19PRE34380842/American Heart Association (AHA)
- CTSA1405/Hydrocephalus Association (HA)
- UO1 HL131003/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- UO1-HL153009/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- K08 HL157653/HL/NHLBI NIH HHS/United States
- U01 HL098147/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

