Multivariable regression models improve accuracy and sensitive grading of antibiotic resistance mutations in Mycobacterium tuberculosis
- PMID: 40032816
- PMCID: PMC11876447
- DOI: 10.1038/s41467-025-57174-1
Multivariable regression models improve accuracy and sensitive grading of antibiotic resistance mutations in Mycobacterium tuberculosis
Abstract
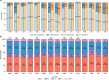
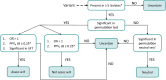
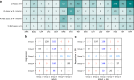
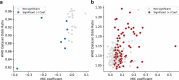
Rapid genotype-based drug susceptibility testing for the Mycobacterium tuberculosis complex (MTBC) relies on a comprehensive knowledgebase of the genetic determinants of resistance. Here we present a catalogue of resistance-associated mutations using a regression-based approach and benchmark it against the 2nd edition of the World Health Organisation (WHO) mutation catalogue. We train multivariate logistic regression models on over 52,000 MTBC isolates to associate binary resistance phenotypes for 15 antitubercular drugs with variants extracted from candidate resistance genes. Regression detects 450/457 (98%) resistance-associated variants identified using the existing method (a.k.a, SOLO method) and grades 221 (29%) more total variants than SOLO. The regression-based catalogue achieves higher sensitivity on average (+3.2 percentage points, pp) than SOLO with smaller average decreases in specificity (-1.0 pp) and positive predictive value (-1.6 pp). Sensitivity gains are highest for ethambutol, clofazimine, streptomycin, and ethionamide as regression graded considerably more resistance-associated variants than SOLO for these drugs. There is no difference between SOLO and regression with regards to meeting the target product profiles set by the WHO for genetic drug susceptibility testing, except for rifampicin, for which regression specificity is below the threshold of 98% at 97%. The regression pipeline also detects isoniazid resistance compensatory mutations in ahpC and variants linked to bedaquiline and aminoglycoside hypersusceptibility. These results inform the continued development of targeted next generation sequencing, whole genome sequencing, and other commercial molecular assays for diagnosing resistance in the MTBC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.C.R. received funding support from FIND through a service contract with UC San Diego. T.C.R. received grant funding from NIH to develop and evaluate a tNGS solution for drug resistant TB (R01AI176401). T.C.R. is a co-founder, board member and unpaid shareholder of Verus Diagnostics Inc. T.C.R. is a co-inventor on patents pertaining to tNGS. The remaining authors declare no competing interests.
Figures
References
-
- WHO. The use of next-generation sequencing for the surveillance of drug-resistant tuberculosis: An implementation manual (World Health Organization, 2023).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

