Synergistic transfer learning and adversarial networks for breast cancer diagnosis: benign vs. invasive classification
- PMID: 40032913
- PMCID: PMC11876678
- DOI: 10.1038/s41598-025-90288-6
Synergistic transfer learning and adversarial networks for breast cancer diagnosis: benign vs. invasive classification
Abstract
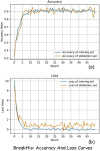
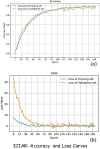
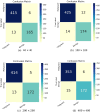
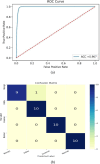
Current breast cancer diagnosis methods often face limitations such as high cost, time consumption, and inter-observer variability. To address these challenges, this research proposes a novel deep learning framework that leverages generative adversarial networks (GANs) for data augmentation and transfer learning to enhance breast cancer classification using convolutional neural networks (CNNs). The framework uses a two-stage augmentation approach. First, a conditional Wasserstein GAN (cWGAN) generates synthetic breast cancer images based on clinical data, enhancing training stability and enabling targeted feature incorporation. Second, traditional augmentation techniques (e.g., rotation, flipping, cropping) are applied to both original and synthetic images. A multi-scale transfer learning technique is also employed, integrating three pre-trained CNNs (DenseNet-201, NasNetMobile, ResNet-101) with a multi-scale feature enrichment scheme, allowing the model to capture features at various scales. The framework was evaluated on the BreakHis dataset, achieving an accuracy of 99.2% for binary classification and 98.5% for multi-class classification, significantly outperforming existing methods. This framework offers a more efficient, cost-effective, and accurate approach for breast cancer diagnosis. Future work will focus on generalizing the framework to clinical datasets and integrating it into diagnostic workflows.
Keywords: Accuracy; Breast cancer diagnosis; Deep learning; Generative adversarial networks (GANs); Transfer learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
-
- Gurcan, M. N., Madabhushi, A. & Rajpoot, N. Pattern recognition in histopathological images: An ICPR 2010 contest. In Recognizing Patterns in Signals, Speech, Images and Videos: ICPR 2010 Contests, Istanbul, Turkey, August 23–26, 2010, Contest Reports 226–234 (Springer, 2010).
-
- Veta, M., Pluim, J. P., Van Diest, P. J. & Viergever, M. A. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng.61(5), 1400–1411 (2014). - PubMed
-
- Robboy, S. J. et al. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch. Pathol. Lab. Med.137(12), 1723–1732 (2013). - PubMed
-
- Pöllänen, I., Braithwaite, B., Haataja, K., Ikonen, T. & Toivanen, P. Current analysis approaches and performance needs for whole slide image processing in breast cancer diagnostics. In 2015 International Conference on Embedded Computer Systems: Architectures, Modeling, and Simulation (SAMOS) 319–325 (IEEE, 2015).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

