Complement activation in tumor microenvironment after neoadjuvant therapy and its impact on pancreatic cancer outcomes
- PMID: 40032924
- PMCID: PMC11876354
- DOI: 10.1038/s41698-025-00848-2
Complement activation in tumor microenvironment after neoadjuvant therapy and its impact on pancreatic cancer outcomes
Abstract
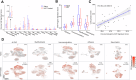
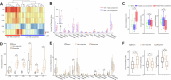
Neoadjuvant therapy (NAT) is increasingly being used for pancreatic ductal adenocarcinoma (PDAC). This study investigates how NAT differentially impacts PDAC's carcinoma cells and the tumor microenvironment (TME). Spatial transcriptomics was used to compare gene expression profiles in carcinoma cells and the TME of 23 NAT-treated versus 13 NAT-naïve PDACs. Findings were validated by single-nucleus RNA sequencing (snRNA-seq) analysis. NAT induces apoptosis and inhibits proliferation of carcinoma cells and coordinately upregulates multiple complement genes (C1R, C1S, C3, C4B and C7) within the TME. Higher TME complement expression following NAT is associated with increased immunomodulatory and neurotrophic cancer-associated fibroblasts (CAFs); more CD4+ T cells; reduced immune exhaustion gene expression, and improved overall survival. snRNA-seq analysis demonstrates C3 complement is mainly upregulated in CAFs. These findings suggest that local complement dynamics could serve as a novel biomarker for prognosis, evaluating treatment response, and guiding therapeutic strategies in NAT-treated PDAC patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Enhanced Complement Expression in the Tumor Microenvironment Following Neoadjuvant Therapy: Implications for Immunomodulation and Survival in Pancreatic Ductal Adenocarcinoma.Res Sq [Preprint]. 2024 May 13:rs.3.rs-4104258. doi: 10.21203/rs.3.rs-4104258/v1. Res Sq. 2024. Update in: NPJ Precis Oncol. 2025 Mar 3;9(1):58. doi: 10.1038/s41698-025-00848-2. PMID: 38798691 Free PMC article. Updated. Preprint.
References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33 (2022). - PubMed
-
- Ferrone, C. R. et al. Pancreatic adenocarcinoma: the actual 5-year survivors. J. Gastrointest. Surg.12, 701–706 (2008). - PubMed
-
- Hu, Z. I. & O’Reilly, E. M. Therapeutic developments in pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol.21, 7–24 (2024). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

