SETD1B-mediated broad H3K4me3 controls proper temporal patterns of gene expression critical for spermatid development
- PMID: 40033033
- PMCID: PMC12012180
- DOI: 10.1038/s41422-025-01080-0
SETD1B-mediated broad H3K4me3 controls proper temporal patterns of gene expression critical for spermatid development
Abstract
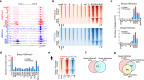
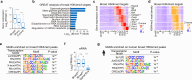
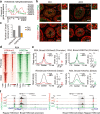
Epigenetic programming governs cell fate determination during development through intricately controlling sequential gene activation and repression. Although H3K4me3 is widely recognized as a hallmark of gene activation, its role in modulating transcription output and timing within a continuously developing system remains poorly understood. In this study, we provide a detailed characterization of the epigenomic landscapes in developing male germ cells. We identified thousands of spermatid-specific broad H3K4me3 domains regulated by the SETD1B-RFX2 axis, representing a previously underappreciated form of H3K4me3. These domains, overlapping with H3K27ac-marked enhancers and promoters, play critical roles in orchestrating robust transcription and accurate temporal control of gene expression. Mechanistically, these broad H3K4me3 compete effectively with regular H3K4me3 for transcriptional machinery, thereby ensuring robust levels and precise timing of master gene expression in mouse spermiogenesis. Disruption of this mechanism compromises the accuracy of transcription dosage and timing, ultimately impairing spermiogenesis. Additionally, we unveil remarkable changes in the distribution of heterochromatin marks, including H3K27me3 and H3K9me2, during the mitosis-to-meiosis transition and completion of meiotic recombination, which closely correlates with gene silencing. This work underscores the highly orchestrated epigenetic regulation in spermatogenesis, highlighting the previously unrecognized role of Setd1b in the formation of broad H3K4me3 domains and transcriptional control, and provides an invaluable resource for future studies toward the elucidation of spermatogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Fei Lan is a scientific co-founder and stockholder of Active Motif Shanghai, Inc. and Alternative Bio, Inc. All other authors declare no competing interests.
Figures




References
-
- Clermont, Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev.52, 198–236 (1972). - PubMed
-
- Soumillon, M. et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep.3, 2179–2190 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- 31930034/National Natural Science Foundation of China (National Science Foundation of China)
- 32100455/National Natural Science Foundation of China (National Science Foundation of China)
- 32170861/National Natural Science Foundation of China (National Science Foundation of China)
- 32288102/National Natural Science Foundation of China (National Science Foundation of China)
- 31925010/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources

