The genomic landscape of gene-level structural variations in Japanese and global soybean Glycine max cultivars
- PMID: 40033060
- PMCID: PMC11985339
- DOI: 10.1038/s41588-025-02113-5
The genomic landscape of gene-level structural variations in Japanese and global soybean Glycine max cultivars
Erratum in
-
Author Correction: The genomic landscape of gene-level structural variations in Japanese and global soybean Glycine max cultivars.Nat Genet. 2025 Jul;57(7):1787. doi: 10.1038/s41588-025-02256-5. Nat Genet. 2025. PMID: 40494993 Free PMC article. No abstract available.
-
Author Correction: The genomic landscape of gene-level structural variations in Japanese and global soybean Glycine max cultivars.Nat Genet. 2025 Oct;57(10):2603. doi: 10.1038/s41588-025-02356-2. Nat Genet. 2025. PMID: 40921790 Free PMC article. No abstract available.
Abstract
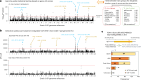
Japanese soybeans are traditionally bred to produce soy foods such as tofu, miso and boiled soybeans. Here, to investigate their distinctive genomic features, including genomic structural variations (SVs), we constructed 11 nanopore-based genome references for Japanese and other soybean lines. Our assembly-based comparative method, designated 'Asm2sv', identified gene-level SVs comprehensively, enabling pangenome analysis of 462 worldwide cultivars and varieties. Based on these, we identified selective sweeps between Japanese and US soybeans, one of which was the pod-shattering resistance gene PDH1. Genome-wide association studies further identified several quantitative trait loci that accounted for large-seed phenotypes of Japanese soybean lines, some of which were also close to regions of the selective sweeps, including PDH1. Notably, specific combinations of alleles, including SVs, were found to increase the seed size of some Japanese landraces. In addition to the differences in cultivation environments, distinct food processing usages might result in changes in Japanese soybean genomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Food & Agriculture Organization of the United Nations. FAOSTAT Statistical Database (FAO, 2021).
-
- Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature463, 178–183 (2010). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

