Proteomic Profiling of Extracellular Vesicles Reveals Potential Biomarkers for Helicobacter pylori Infection and Gastric Cancer
- PMID: 40033163
- PMCID: PMC11876490
- DOI: 10.1111/hel.70022
Proteomic Profiling of Extracellular Vesicles Reveals Potential Biomarkers for Helicobacter pylori Infection and Gastric Cancer
Abstract
Background: Helicobacter pylori (H. pylori) has been identified as a type I carcinogen and contributes to a high rate of gastric cancer (GC), especially in Eastern Asia. Extracellular vesicles (EVs) have the potential to be used to detect various cancer types and diseases. However, the protein markers in EVs for the prognosis of H. pylori infection and GC are unknown. We aim to identify the proteins within EVs derived from a gastric epithelial cell line (AGS) infected with H. pylori by using LC-MS/MS.
Materials and methods: EVs were isolated from AGS cells infected with high- and low-virulence H. pylori (strains TN2wt and Tx30a) by ultracentrifugation. Proteins within these EVs were identified and analyzed for potential marker candidates through bioinformatics. Proteins in H. pylori-derived EVs (HpEVs) from bacterial culture supernatant and HpEVs derived from H. pylori-infected AGS cells were elucidated.
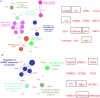
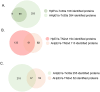
Results: Differentially expressed proteins by proteomic analysis in AGSEVs-Tx30a vs. AGSEVs-noninfected (NI) and AGSEVs-TN2wt vs. AGSEVs-NI were 107 and 55 proteins, respectively. Bioinformatics of these proteomes revealed that essential proteins for H. pylori survival and pathogenicity including outer membrane proteins, metabolism-related, host cell infection-related, and virulence-related proteins were observed in HpEVs. Interestingly, EVs derived from AGS cells infected with H. pylori TN2wt significantly contained multiple proteins related to GC (ATP6V0A1, GAPDH, HINT1, LYZ, and RBX1).
Conclusion: This study provides a comprehensive protein profile of EVs from H. pylori-infected AGS cells and HpEVs, which could serve as liquid-based biomarkers in the future for screening H. pylori infection, especially GC-related.
Keywords: Helicobacter pylori; biomarker; extracellular vesicles; gastric cancer; proteomics.
© 2025 The Author(s). Helicobacter published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Hooi J. K. Y., Lai W. Y., Ng W. K., et al., “Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta‐Analysis,” Gastroenterology 153, no. 2 (2017): 420–429. - PubMed
-
- Suerbaum S. and Michetti P., “ Helicobacter pylori Infection,” New England Journal of Medicine 347, no. 15 (2002): 1175–1186. - PubMed
-
- Peek R. M. and Blaser M. J., “ Helicobacter pylori and Gastrointestinal Tract Adenocarcinomas,” Nature Reviews Cancer 2, no. 1 (2002): 28–37. - PubMed
-
- Bray F., Laversanne M., Sung H., et al., “Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries,” CA: A Cancer Journal for Clinicians 74, no. 3 (2024): 229–263. - PubMed
-
- Correa P., “Human Gastric Carcinogenesis: A Multistep and Multifactorial Process‐First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention,” Cancer Research 52, no. 24 (1992): 6735–6740. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

