The role of senescence-related genes in major depressive disorder: insights from machine learning and single cell analysis
- PMID: 40033248
- PMCID: PMC11874787
- DOI: 10.1186/s12888-025-06542-8
The role of senescence-related genes in major depressive disorder: insights from machine learning and single cell analysis
Abstract
Background: Evidence indicates that patients with Major Depressive Disorder (MDD) exhibit a senescence phenotype or an increased susceptibility to premature senescence. However, the relationship between senescence-related genes (SRGs) and MDD remains underexplored.
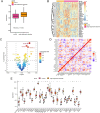
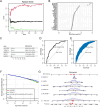
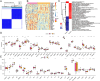
Methods: We analyzed 144 MDD samples and 72 healthy controls from the GEO database to compare SRGs expression. Using Random Forest (RF) and Support Vector Machine-Recursive Feature Elimination (SVM-RFE), we identified five hub SRGs to construct a logistic regression model. Consensus cluster analysis, based on SRGs expression patterns, identified subclusters of MDD patients. Weighted Gene Co-expression Network Analysis (WGCNA) identified gene modules strongly linked to each cluster. Single-cell RNA sequencing was used to analyze MDD SRGs functions.
Results: The five hub SRGs: ALOX15B, TNFSF13, MARCH 15, UBTD1, and MAPK14 showed differential expression between MDD patients and controls. Diagnostics models based on these hub genes demonstrated high accuracy. The hub SRGs correlated positively with neutrophils and negatively with T lymphocytes. SRGs expression pattern revealed two distinct MDD subclusters. WGCNA identified significant gene modules within these subclusters. Additionally, individual endothelial cells with high senescence scores were found to interact with astrocytes via the Notch signaling pathway, suggesting a specific role in MDD pathogenesis.
Conclusion: This comprehensive study elucidates the significant role of SRGs in MDD, highlighting the importance of the Notch signaling pathway in mediating senescence effects.
Keywords: Bulk RNA analysis; Endothelial cells; Major depressive disorder; Senescence; Single-cell RNA analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The patients involved in the GEO database have obtained ethical approval. Our study is based on open data, there are no ethical issues and other conflicts of interest. Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

