Functional validation to explore the protective role of miR-223 in Staphylococcus aureus-induced bovine mastitis
- PMID: 40033327
- PMCID: PMC11877765
- DOI: 10.1186/s40104-025-01152-6
Functional validation to explore the protective role of miR-223 in Staphylococcus aureus-induced bovine mastitis
Abstract
Background: Mastitis caused by Staphylococcus aureus (S. aureus) is one of the most intractable problems for the dairy industry, causing significantly reduced milk yields and early slaughter of cows worldwide. MicroRNAs (miRNAs) can post-transcriptionally regulate gene expression and studies in recent years have shown the importance of miRNA-associated gene regulation in S. aureus-induced mastitis.
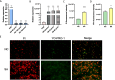
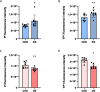
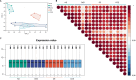
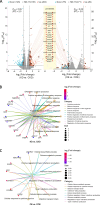
Results: In this study, to investigate the role of miR-223 in mastitis, we performed experiments to overexpress and suppress miR-223 in an immortalized bovine mammary epithelial cell line (MAC-T) infected with S. aureus. Overexpression of miR-223 in MAC-T cells repressed cell apoptosis and necrosis induced by S. aureus infection, whereas suppression of miR-223 had the opposite effect. Transcriptome expression profiling with weighted gene co-expression network analysis (WGCNA) and gene set variation analysis (GSVA) showed that miR-223 affects apoptosis and inflammation-related pathways. Furthermore, differentially expressed (DE) genes were evaluated, and genes exhibiting contrasting expression trends in the miR-223 overexpressed and suppressed groups were assessed as potential target genes of miR-223. Potential target genes, including CDC25B, PTPRF, DCTN1, and DPP9, were observed to be associated with apoptosis and necroptosis. Finally, through integrative analysis of genome-wide association study (GWAS) data and the animal quantitative trait loci (QTL) database, we determined that target genes of miR-223 were significantly enriched in single-nucleotide polymorphisms (SNP) and QTLs related to somatic cell count (SCC) and mastitis.
Conclusion: In summary, miR-223 has an inhibitory effect on S. aureus-induced cell apoptosis and necrosis by regulating PTPRF, DCTN1, and DPP9. These genes were significantly enriched in QTL regions associated with bovine mastitis resistance, underscoring their relevance in genetic regulation of disease resilience. Our findings provide critical genetic markers for enhancing mastitis resistance, particularly S. aureus-induced mastitis, through selective breeding. This work offers valuable insights for developing cattle with improved resistance to mastitis via targeted genetic selection.
Keywords: Staphylococcus aureus; Bovine mastitis; Gene regulation; Mammary epithelial cells; MiR-223.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The cell line MAC-T used in this study was obtained from primary bovine mammary alveolar cells [18], and all experiments were conducted in accordance with relevant guidelines and regulations. The S. aureus strain used was selected from 191 S. aureus strains, which were originally isolated from milk samples of 1,112 lactating Holstein cows [63], and all procedures involving bacterial handling and experimentation were conducted in compliance with biosafety guidelines approved by China Agricultural University. No human or animal subjects were directly involved in this research; therefore, specific ethical approval and consent to participate were not required. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures




References
-
- De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. Invited review: Mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J Dairy Sci. 2012;95:1025–40. 10.3168/jds.2010-4074. - PubMed
-
- Gonçalves JL, Kamphuis C, Martins CMMR, Barreiro JR, Tomazi T, Gameiro AH, et al. Bovine subclinical mastitis reduces milk yield and economic return. Livest Sci. 2018;210:25–32. 10.1016/j.livsci.2018.01.016.
-
- Kerro Dego O, van Dijk JE, Nederbragt H. Factors involved in the early pathogenesis of bovine Staphylococcus aureus mastitis with emphasis on bacterial adhesion and invasion. Vet Q. 2002;24:181–98. 10.1080/01652176.2002.9695135. - PubMed
-
- Barkema HW, von Keyserlingk MA, Kastelic JP, Lam TJ, Luby C, Roy JP, et al. Invited review: Changes in the dairy industry affecting dairy cattle health and welfare. J Dairy Sci. 2015;98:7426–45. 10.3168/jds.2015-9377. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

