An international observational study validating gene-expression sepsis immune subgroups
- PMID: 40033354
- PMCID: PMC11877781
- DOI: 10.1186/s13054-025-05319-5
An international observational study validating gene-expression sepsis immune subgroups
Abstract
Background: Sepsis gene-expression sub-phenotypes with prognostic and theranostic potential have been discovered. These have been identified retrospectively and have not been translated to methods that could be deployed at the bedside. We aimed to identify subgroups of septic patients at high-risk of poor outcome, using a rapid, multiplex RNA-based test.
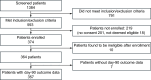
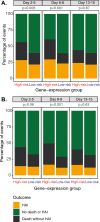
Methods: Adults with sepsis, in the intensive care unit (ICU) were recruited from 17 sites in the United Kingdom, Sweden and France. Blood was collected at days 2-5 (S1), 6-8 (S2) and 13-15 (S3) after ICU admission and analyzed centrally. Patients were assigned into 'high' and 'low' risk groups using two models previously developed for the Immune-Profiling Panel prototype on the bioMérieux FilmArray® system.
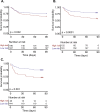
Results: 357 patients were recruited (March 2021-November 2022). 69% were male with a median age of 67 years, APACHE II score of 21 and a 30% 90-day mortality rate. The proportions of high-risk patients decreased over the three sampling times (model 1: 53%, 40%, 15% and model 2: 81%, 74%, 37%). In model 1, 90-day mortality was higher in a high-risk group at each time (S1: 35% vs 24%, p = 0.04; S2: 43% vs 20%, p < 0.001; S3: 52% vs 24%, p = 0.007). In model 2, mortality was only significantly different at the second sampling time (S1: 30% vs 27%, p = 0.77; S2: 34% vs 14%, p = 0.002; S3: 35% vs 23%, p = 0.13).
Conclusions: Gene-expression diagnostics can identify patients with sepsis at high-risk of poor outcomes and could be used to identify patients for precision medicine trials.
Registration: ISRCTN11364482 Registered 24th September 2020.
Keywords: Gene-expression; Prospective study; Sepsis; Transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study gained ethical approval in each participating country: The Health and Care Research Wales research ethics committee (20/LO/1163) in the UK; the Comité de Protection des Personnes Ile de France X in France and the Swedish Ethical Review Authority in Sweden. Written informed consent was provided by participants or their legal representatives following local procedures. Competing Interests: ACG reports that outside of this work he has received consulting fees from AstraZeneca, Beckman Coulter, and VVB Bio, speaker fees from Fresenius Kabi and grant support from the NIHR HTA programme. DA reports grant support from the NIHR for work outside this project. FP reports that he has received speaker and consulting fees from Gilead outside of this work. JFT reported consulting and lecture fees for bioMerieux unrelated to the submitted work, consulting fees for Merck, Pfizer, Advanz pharma, Menarini, and lecture fees from Merck, Pfizer, Gilead, Mundipharma Qiagen unrelated to the article. JFT also declared research grants to his research group Merck, Pfizer. NP has served as an advisor or speaker for Moderna and AstraZeneca. EP, SB, AF and JFL are bioMerieux employees. EP and SB are co-inventors of patents related to the topic of the manuscript. GV has received research grants form bioMérieux and SOS Oxygène outside of this work, and support for attending meeting from SOS Oxygène and Oxyvie. DB reports lecture fees for bioMérieux unrelated to the submitted work and consulting for Synairgen and Abioinc. JDR has received speaker fees from Fisher&Paykel. TJ-F has received consulting/speaker fees from MSD, Menarini, bioMérieux, Qiagen, Shionogi, Mundipharma and Pfizer. A-CL reports support from bioMérieux for work outside of this project. NDP reports grants, consulting and speaker fees from AstraZeneca and speaker fees from Moderna. MS reports grants from Gentin, consulting fees from Biotest, Matisse, deePull, Volition, Hemotune and Sanofi and speaker fees from bioMérieux and AOP. The authors Dr Antcliffe and Prof Gordon are affiliated with the Department of Health and Social Care, Centre for Antimicrobial Optimization at Imperial College, London. Other authors declare that they have no competing interests.
Figures
References
-
- Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20(4):195–203. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

