STModule: identifying tissue modules to uncover spatial components and characteristics of transcriptomic landscapes
- PMID: 40033360
- PMCID: PMC11874447
- DOI: 10.1186/s13073-025-01441-9
STModule: identifying tissue modules to uncover spatial components and characteristics of transcriptomic landscapes
Abstract
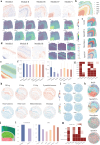
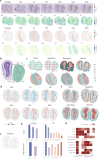
Here we present STModule, a Bayesian method developed to identify tissue modules from spatially resolved transcriptomics that reveal spatial components and essential characteristics of tissues. STModule uncovers diverse expression signals in transcriptomic landscapes such as cancer, intraepithelial neoplasia, immune infiltration, outcome-related molecular features and various cell types, which facilitate downstream analysis and provide insights into tumor microenvironments, disease mechanisms, treatment development, and histological organization of tissues. STModule captures a broader spectrum of biological signals compared to other methods and detects novel spatial components. The tissue modules characterized by gene sets demonstrate greater robustness and transferability across different biopsies. STModule: https://github.com/rwang-z/STModule.git .
Keywords: Bayesian model; Spatial expression components; Spatially resolved transcriptomics; Tissue module.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Moncada R, Barkley D, Wagner F, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38(3):333–42. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

