Lysine 2-hydroxyisobutyrylation of HXK1 alters energy metabolism and KATP channel function in the atrium from patients with atrial fibrillation
- PMID: 40033384
- PMCID: PMC11874433
- DOI: 10.1186/s12964-025-02108-z
Lysine 2-hydroxyisobutyrylation of HXK1 alters energy metabolism and KATP channel function in the atrium from patients with atrial fibrillation
Abstract
Background: Atrial fibrillation (AF) is the most common form of arrhythmia and is a growing clinical problem. Post-translational modifications (PTMs) constitute crucial epigenetic mechanisms but modification of lysine 2-hydroxyisobutyrylation (Khib) in AF is still unknown. This study aimed to investigate the role and mechanism of Khib in AF.
Methods: PTM proteomics was applied in the human atrial tissue from AF and sinus rhythm patients with heart valve disease during cardiac surgery to identify the Khib sites. The functional changes of differential modification sites were further validated at the cellular level. Cellular electrophysiology was performed to record the ion channel current and action potential duration (APD).
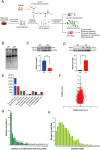
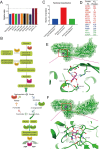
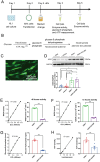
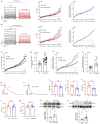
Results: The modification of 124 Khib sites in 35 proteins and 67 sites in 48 proteins exhibited significant increase or decrease in AF compared to sinus rhythm. Ten Khib sites were included in energy metabolism-related signaling pathways (HXK1, TPIS, PGM1, and ODPX in glycolysis; MDHC and IDH3A in tricarboxylic acid cycle; NDUS2, ETFB, ADT3, and ATPB in oxidative respiratory chain). Importantly, decreased HXK1 K418hib regulated by HDAC2 attenuated the original chemical binding domain between HXK1 and glucose, inhibited the binding ability between HXK1 and glucose, and reduced catalytic ability of the enzyme, resulting in low production of glucose-6-phosphate and ATP. Further, it also increased Kir6.2 protein and the current of KATP channel, and decreased APD.
Conclusions: This study demonstrates the importance of Khib to catalysis of HXK1 and reveals molecular mechanisms of HXK1 K418hib in AF, providing new insight into strategies of AF.
Keywords: 2-hydroxyisobutyrylation; Acylation; Atrial fibrillation; Catalysis; Post-translational modification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer.Front Oncol. 2022 Sep 30;12:1001807. doi: 10.3389/fonc.2022.1001807. eCollection 2022. Front Oncol. 2022. PMID: 36249039 Free PMC article.
-
Salt stress downregulates 2-hydroxybutyrylation in Arabidopsis siliques.J Proteomics. 2022 Jan 6;250:104383. doi: 10.1016/j.jprot.2021.104383. Epub 2021 Sep 22. J Proteomics. 2022. PMID: 34562664
-
Proteomic analysis of protein lysine 2-hydroxyisobutyrylation (Khib) in soybean leaves.BMC Plant Biol. 2023 Jan 12;23(1):23. doi: 10.1186/s12870-022-04033-6. BMC Plant Biol. 2023. PMID: 36631736 Free PMC article.
-
Functions of lysine 2-hydroxyisobutyrylation and future perspectives on plants.Proteomics. 2023 Oct;23(19):e2300045. doi: 10.1002/pmic.202300045. Epub 2023 Jun 20. Proteomics. 2023. PMID: 37338329 Review.
-
Research advances in protein lysine 2-hydroxyisobutyrylation: From mechanistic regulation to disease relevance.J Cell Physiol. 2024 Dec;239(12):e31435. doi: 10.1002/jcp.31435. Epub 2024 Oct 1. J Cell Physiol. 2024. PMID: 39351825 Review.
Cited by
-
Post-translational acylation of proteins in cardiac hypertrophy.Nat Rev Cardiol. 2025 Apr 14. doi: 10.1038/s41569-025-01150-1. Online ahead of print. Nat Rev Cardiol. 2025. PMID: 40229510 Review.
References
-
- Nattel S, Atrial Fibrillation and Body Composition. Is it fat or lean that ultimately determines the risk?? J Am Coll Cardiol. 2017;69:2498–501. - PubMed
-
- Kirchhof P, Benussi S, Kotecha D, Ahlsson A, Atar D, Casadei B, Castella M, Diener HC, Heidbuchel H, Hendriks J, et al. 2016 ESC guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur Heart J. 2016;37:2893–962. - PubMed
-
- Zhang D, Wu CT, Qi X, Meijering RA, Hoogstra-Berends F, Tadevosyan A, Cubukcuoglu Deniz G, Durdu S, Akar AR, Sibon OC, et al. Activation of histone deacetylase-6 induces contractile dysfunction through derailment of α-tubulin proteostasis in experimental and human atrial fibrillation. Circulation. 2014;129:346–58. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

