Genome-wide CRISPR/Cas9 screening identifies key profibrotic regulators of TGF-β1-induced epithelial-mesenchymal transformation and pulmonary fibrosis
- PMID: 40034336
- PMCID: PMC11872725
- DOI: 10.3389/fmolb.2025.1507163
Genome-wide CRISPR/Cas9 screening identifies key profibrotic regulators of TGF-β1-induced epithelial-mesenchymal transformation and pulmonary fibrosis
Abstract
Background: The idiopathic pulmonary fibrosis (IPF) is a progressive and lethal interstitial lung disease with high morbidity and mortality. IPF is characterized by excessive extracellular matrix accumulation (ECM) and epithelial-mesenchymal transformation (EMT). To date, few anti-fibrotic therapeutics are available to reverse the progression of pulmonary fibrosis, and it is important to explore new profibrotic molecular regulators mediating EMT and pulmonary fibrosis.
Methods: Based on our model of TGF-β1-induced EMT in BEAS-2B cells, we performed the genome-wide CRISPR/Cas9 knockout (GeCKO) screening technique, pathway and functional enrichment analysis, loss-of-function experiment, as well as other experimental techniques to comprehensively investigate profibrotic regulators contributing to EMT and the pathogenesis of pulmonary fibrosis.
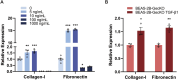
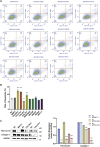
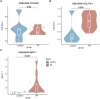
Results: Utilizing the GeCKO library screening, we identified 76 top molecular regulators. Ten candidate genes were subsequently confirmed by integrating the high-throughput data with findings from pathway and functional enrichment analysis. Among the candidate genes, knockout of COL20A1 and COL27A1 led to decreased mRNA expression of ECM components (Fibronectin and Collagen-I), as well as an increased rate of cell apoptosis. The mRNA expression of Collagen-I, together with the cell viability and migration, were inhibited when knocking out the WNT11. In addition, a decrease in the protein deposition of ECM components was observed by suppressing the expression of COL20A1, COL27A1, and WNT11.
Conclusion: Our study demonstrates that the COL20A1, COL27A1, and WNT11 serve as key profibrotic regulators of EMT. Gaining understanding and insights into these key profibrotic regulators of EMT paves the way for the discovery of new therapeutic targets against the onset and progression of IPF.
Keywords: COL20A1; COL27A1; TGF-β1; WNT11; epithelial-mesenchymal transformation; extracellular matrix; genome-wide CRISPR/Cas9 knockout screening; idiopathic pulmonary fibrosis.
Copyright © 2025 Tan, Wang, Ye, Kasimu, Li, Luo, Yi and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Antoniou K., Markopoulou K., Tzouvelekis A., Trachalaki A., Vasarmidi E., Organtzis J., et al. (2020). Efficacy and safety of nintedanib in a Greek multicentre idiopathic pulmonary fibrosis registry: a retrospective, observational, cohort study. ERJ Open Res. 6 (1). 10.1183/23120541.00172-2019 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

