Integrating Radiosensitivity Index and Radiation Resistance Related Index Improves Prostate Cancer Outcome Prediction
- PMID: 40034476
- PMCID: PMC11872654
- DOI: 10.1016/j.adro.2025.101713
Integrating Radiosensitivity Index and Radiation Resistance Related Index Improves Prostate Cancer Outcome Prediction
Erratum in
-
Erratum to 'Integrating Radiosensitivity Index and Radiation Resistance Related Index Improves Prostate Cancer Outcome Prediction' [Advances in Radiation Oncology (2025) 10, 101713].Adv Radiat Oncol. 2025 Aug 14;10(8):101830. doi: 10.1016/j.adro.2025.101830. eCollection 2025 Aug. Adv Radiat Oncol. 2025. PMID: 40842713 Free PMC article.
Abstract
Purpose: This study aimed to establish a nomogram combining 31-gene signature (31-GS), radiosensitivity index (RSI), and radiation-resistance-related gene index (RRRI) to predict recurrence in prostate cancer (PCa) patients.
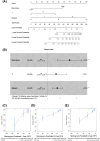
Methods and materials: Transcriptome data of PCa were obtained from gene expression omnibus and the cancer genome atlas to validate the predictive potential of 3 sets of published biomarkers, namely, 31-GS, RSI, and RRRI. To adjust these markers for the characteristics of PCa, we analyzed 4 PCa-associated radiosensitivity predictive indices based on 31-GS, RSI, and RRRI by the Cox analysis and least absolute shrinkage and selection operator regression analysis. Time-dependent receiver operating characteristic curves, decision curve analyses, integrated discrimination improvement, net reclassification improvement and decision tree model construction were used to compare the radiosensitivity predictive ability of these 4 gene signatures. Key modules and associated functions were identified through a weighted gene co-expression network analysis and gene function enrichment analysis. A nomogram was built to improve the recurrence-prediction capability.
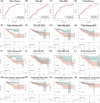
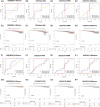
Results: We validated and compared the predictive potential of 2 published predictive indices. Based on the 31-GS, RSI, and RRRI, we analyzed 4 PCa-associated radiosensitivity predictive indices: 14Genes, RSI, RRRI, and 20Genes. Among them, 14Genes showed the most promising predictive performance and discriminative capacity. Genes in the key module defined by the 14Genes model were significantly enriched in radiation therapy-related cell death pathways. The area under receiver operating characteristic curve and decision tree variable importance for 14Genes was the highest in the cancer genome atlas and Gene Expression Omnibus Series (GSE) cohorts.
Conclusions: This study successfully established a radiosensitivity-related nomogram, which had excellent performance in predicting recurrence in patients with PCa. For patients who received radiation therapy, the 20Genes and RRRI models can be used to predict recurrence-free survival, whereas 20Genes is more radiation therapy-specific but needs further external validation.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. - PubMed
-
- Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol. 2016;17(11):1612–1620. - PubMed
LinkOut - more resources
Full Text Sources

