This is a preprint.
Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms
- PMID: 40034650
- PMCID: PMC11875243
- DOI: 10.1101/2025.02.25.640204
Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms
Abstract
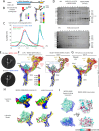
Microtubule (MT) branch nucleation is fundamental for building parallel MT networks in eukaryotic cells. In plants and metazoans, MT branch nucleation requires Augmin and NEDD1 proteins which bind along MTs and then recruit and activate the gamma-tubulin ring complex (γ-TuRC). Augmin is a fork-shaped assembly composed of eight coiled-coil subunits, while NEDD1 is a WD40 β-propellor protein that bridges across MTs, Augmin, and γ-TuRC during MT branch nucleation. Here, we reconstitute hetero-tetrameric and hetero-octameric Arabidopsis thaliana Augmin assemblies, resolve their subunit interactions using crosslinking mass spectrometry and determine 3.7 to 7.3-Å cryo-EM structures for the V-junction and extended regions of Augmin. These structures allowed us to generate a complete de novo plant Augmin model that reveals the long-range multi coiled-coil interfaces that stabilize its 40-nm hetero-octameric fork-shaped organization. We discovered the dual calponin homology (CH) domain forming its MT binding site at the end of its V-junction undertake open and closed conformations. We determined a 12-Å dimeric Augmin cryo-EM structure revealing Augmin undergoes anti-parallel dimerization through two conserved surfaces along Augmin's extended region. We reconstituted the NEDD1 WD40 β-propellor with Augmin revealing it directly binds on top its V-junction and enhances Augmin dimerization. Our studies suggest that cooperativity between the Augmin dual CH domains and NEDD1 WD40 binding site may regulate Augmin V-junction dual binding to MT lattices. This unique V-shaped dual binding and organization anchors Augmins along MTs generating a platform to recruit γ-TuRC and activate branched MT nucleation.
Keywords: AUG; Augmin; Cryo-EM; Haus; NEDD1; branch nucleation; gamma-tubulin ring complex; microtubule.
Figures






Similar articles
-
Cross-linking mass spectrometry identifies new interfaces of Augmin required to localise the γ-tubulin ring complex to the mitotic spindle.Biol Open. 2017 May 15;6(5):654-663. doi: 10.1242/bio.022905. Biol Open. 2017. PMID: 28351835 Free PMC article.
-
Molecular architecture of the augmin complex.Nat Commun. 2022 Sep 16;13(1):5449. doi: 10.1038/s41467-022-33227-7. Nat Commun. 2022. PMID: 36114186 Free PMC article.
-
Structure of the microtubule anchoring factor NEDD1 bound to the γ-tubulin ring complex.bioRxiv [Preprint]. 2024 Nov 5:2024.11.05.622067. doi: 10.1101/2024.11.05.622067. bioRxiv. 2024. Update in: J Cell Biol. 2025 Aug 4;224(8):e202410206. doi: 10.1083/jcb.202410206. PMID: 39574704 Free PMC article. Updated. Preprint.
-
Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells.New Phytol. 2019 Jun;222(4):1705-1718. doi: 10.1111/nph.15705. Epub 2019 Feb 25. New Phytol. 2019. PMID: 30681146 Review.
-
Microtubule organization defects in Arabidopsis thaliana.Plant Biol (Stuttg). 2020 Nov;22(6):971-980. doi: 10.1111/plb.13114. Epub 2020 Aug 26. Plant Biol (Stuttg). 2020. PMID: 32215997 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
