This is a preprint.
Assessment of MYC Gene and WNT Pathway Alterations in Early-Onset Colorectal Cancer Among Hispanic/Latino Patients Using Integrated Multi-Omics Approaches
- PMID: 40034762
- PMCID: PMC11875251
- DOI: 10.1101/2024.12.05.24318588
Assessment of MYC Gene and WNT Pathway Alterations in Early-Onset Colorectal Cancer Among Hispanic/Latino Patients Using Integrated Multi-Omics Approaches
Abstract
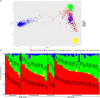
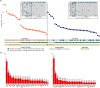
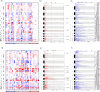
Colorectal cancer (CRC) has increased at an alarming rate amongst younger (< 50 years) individuals. Such early-onset colorectal cancer (EOCRC) has been particularly notable within the Hispanic/Latino population. Yet, this population has not been sufficiently profiled in terms of two critical elements of CRC -- the MYC proto-oncogene and WNT signaling pathway. Here, we performed a comprehensive multi-omics analysis on 30 early-onset and 37 late-onset CRC (≥ 50 years) samples from Hispanic/Latino patients. Our analysis included DNA exome sequencing for somatic mutations, somatic copy number alterations, and global and local genetic similarity. Using RNA sequencing, we also assessed differential gene expression, cellular pathways, and gene fusions. We then compared our findings from early-onset Hispanic/Latino patient samples with publicly available data from Non-Hispanic White cohorts. Across all early-onset patients, which had a median 1000 Genomes Project Peruvian-in-Lima-like (1KG-PEL-like) genetic similarity proportion of 60%, we identified 41 WNT pathway genes with significant mutations. Six important examples were APC, TCF7L2, DKK1, DKK2, FZD10, and LRP5. Notably, patients with mutations in DKK1 and DKK2 had the highest 1KG-PEL-like proportion (79%). When we compared the Hispanic/Latino cohort to the Non-Hispanic White cohorts, four of these key genes -- DKK1, DKK2, FZD10, and LRP5 -- were significant in both risk association analyses and differential gene expression. Interestingly, early-onset tumors (vs. late-onset) exhibited distinct somatic copy number alterations and gene expression profiles; the differences included MYC and drug-targetable WNT pathway genes. We also identified a novel WNT gene fusion, RSPO3, in early-onset tumors; it was associated with enhanced WNT signaling. This integrative analysis underscores the distinct molecular features of EOCRC cancer in the Hispanic/Latino population; reveals potential avenues for tailored precision medicine therapies; and emphasizes the importance of multi-omics approaches in studying colorectal carcinogenesis. We expect this data to help contribute towards reducing cancer health disparities.
Significance: This study offers multi-omics profiling analysis of early-onset colorectal cancer (EOCRC) in an underserved community, explores the implications of MYC gene and WNT pathway alterations, and provides critical insights into cancer health disparities.
Figures


References
-
- Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, Mirlacher M, Tapia C, Simon R, Sauter G, Terracciano L, Tornillo L. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007. Jul;60(7):768–72. doi: 10.1136/jcp.2006.038281. Epub 2006 Aug 1. - DOI - PMC - PubMed
-
- Benjamin D, Sato T, Cibulskis K, et al. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 861054; doi: 10.1101/861054. - DOI
-
- Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010. Nov 12;5(11):e13978. doi: 10.1371/journal.pone.0013978. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
