Large-scale phenotypic and genomic analysis of Listeria monocytogenes reveals diversity in the sensitivity to quaternary ammonium compounds but not to peracetic acid
- PMID: 40035557
- PMCID: PMC12016499
- DOI: 10.1128/aem.01829-24
Large-scale phenotypic and genomic analysis of Listeria monocytogenes reveals diversity in the sensitivity to quaternary ammonium compounds but not to peracetic acid
Erratum in
-
Erratum for Ivanova et al., "Large-scale phenotypic and genomic analysis of Listeria monocytogenes reveals diversity in the sensitivity to quaternary ammonium compounds but not to peracetic acid".Appl Environ Microbiol. 2025 Jul 23;91(7):e0095625. doi: 10.1128/aem.00956-25. Epub 2025 Jun 3. Appl Environ Microbiol. 2025. PMID: 40459295 Free PMC article. No abstract available.
Abstract
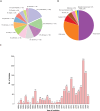
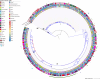
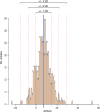
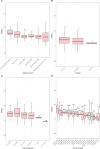
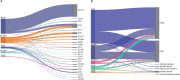
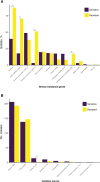
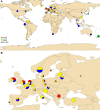
Listeria monocytogenes presents a significant concern for the food industry due to its ability to persist in the food processing environment. One of the factors contributing to its persistence is decreased sensitivity to disinfectants. Our objective was to assess the diversity of L. monocytogenes sensitivity to food industry disinfectants by testing the response of 1,671 L. monocytogenes isolates to quaternary ammonium compounds (QACs) and 414 isolates to peracetic acid (PAA) using broth microdilution and growth curve analysis assays, respectively, and to categorize the isolates into sensitive and tolerant. A high phenotype-genotype concordance (95%) regarding tolerance to QACs was obtained by screening the genomes for the presence of QAC tolerance-associated genes bcrABC, emrE, emrC, and qacH. Based on this high concordance, we assessed the QAC genes' dissemination among publicly available L. monocytogenes genomes (n = 39,196). Overall, QAC genes were found in 23% and 28% of the L. monocytogenes collection in this study and in the global data set, respectively. bcrABC and qacH were the most prevalent genes, with bcrABC being the most detected QAC gene in the USA, while qacH dominated in Europe. No significant differences (P > 0.05) in the PAA tolerance were detected among isolates belonging to different lineages, serogroups, clonal complexes, or isolation sources, highlighting limited variation in the L. monocytogenes sensitivity to this disinfectant. The present work represents the largest testing of L. monocytogenes sensitivity to important food industry disinfectants at the phenotypic and genomic level, revealing diversity in the tolerance to QACs while all isolates showed similar sensitivity to PAA.
Importance: Contamination of Listeria monocytogenes within food processing environments is of great concern to the food industry due to challenges in eradicating the isolates once they become established and persistent in the environment. Genetic markers associated with increased tolerance to certain disinfectants have been identified, which alongside other biotic and abiotic factors can favor the persistence of L. monocytogenes in the food production environment. By employing a comprehensive large-scale phenotypic testing and genomic analysis, this study significantly enhances the understanding of the L. monocytogenes tolerance to quaternary ammonium compounds (QACs) and the genetic determinants associated with the increased tolerance. We provide a global overview of the QAC genes prevalence among public L. monocytogenes sequences and their distribution among clonal complexes, isolation sources, and geographical locations. Additionally, our comprehensive screening of the peracetic acid (PAA) sensitivity shows that this disinfectant can be used in the food industry as the lack of variation in sensitivity indicates reliable effect and no apparent possibility for the emergence of tolerance.
Keywords: Listeria monocytogenes; disinfectants; food industry; peracetic acid; quaternary ammonium compounds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry.Int J Food Microbiol. 2017 Jan 16;241:215-224. doi: 10.1016/j.ijfoodmicro.2016.10.025. Epub 2016 Oct 21. Int J Food Microbiol. 2017. PMID: 27810443
-
Genomic analysis of Listeria monocytogenes from US food processing environments reveals a high prevalence of QAC efflux genes but limited evidence of their contribution to environmental persistence.BMC Genomics. 2022 Jul 4;23(1):488. doi: 10.1186/s12864-022-08695-2. BMC Genomics. 2022. PMID: 35787787 Free PMC article.
-
Tolerance of Listeria monocytogenes to Quaternary Ammonium Sanitizers Is Mediated by a Novel Efflux Pump Encoded by emrE.Appl Environ Microbiol. 2015 Nov 20;82(3):939-53. doi: 10.1128/AEM.03741-15. Print 2016 Feb 1. Appl Environ Microbiol. 2015. PMID: 26590290 Free PMC article.
-
Biocide use in the food industry and the disinfectant resistance of persistent strains of Listeria monocytogenes and Escherichia coli.Symp Ser Soc Appl Microbiol. 2002;(31):111S-120S. Symp Ser Soc Appl Microbiol. 2002. PMID: 12481836 Review.
-
Potential Impact of the Resistance to Quaternary Ammonium Disinfectants on the Persistence of Listeria monocytogenes in Food Processing Environments.Front Microbiol. 2016 May 2;7:638. doi: 10.3389/fmicb.2016.00638. eCollection 2016. Front Microbiol. 2016. PMID: 27199964 Free PMC article. Review.
Cited by
-
Early detection and population dynamics of Listeria monocytogenes in naturally contaminated drains from a meat processing plant.Front Microbiol. 2025 Apr 9;16:1541481. doi: 10.3389/fmicb.2025.1541481. eCollection 2025. Front Microbiol. 2025. PMID: 40270812 Free PMC article.
-
Repeated biocide treatments cause changes to the microbiome of a food industry floor drain biofilm model.Front Microbiol. 2025 Mar 14;16:1542193. doi: 10.3389/fmicb.2025.1542193. eCollection 2025. Front Microbiol. 2025. PMID: 40160267 Free PMC article.
References
-
- Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Girones R, Herman L, Koutsoumanis K, Nørrung B, et al. 2018. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J 16:e05134. doi: 10.2903/j.efsa.2018.5134 - DOI - PMC - PubMed
-
- Belias A, Sullivan G, Wiedmann M, Ivanek R. 2022. Factors that contribute to persistent Listeria in food processing facilities and relevant interventions: a rapid review. Food Control 133:108579. doi: 10.1016/j.foodcont.2021.108579 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

