Characterization of group I introns in generating circular RNAs as vaccines
- PMID: 40036878
- PMCID: PMC11879131
- DOI: 10.1093/nar/gkaf089
Characterization of group I introns in generating circular RNAs as vaccines
Abstract
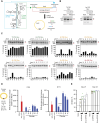
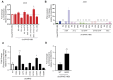
Circular RNAs are an increasingly important class of RNA molecules that can be engineered as RNA vaccines and therapeutics. Here, we screened eight different group I introns for their ability to circularize and delineated different features that are important for their function. First, we identified the Scytalidium dimidiatum group I intron as causing minimal innate immune activation inside cells, underscoring its potential to serve as an effective RNA vaccine without triggering unwanted reactogenicity. Additionally, mechanistic RNA structure analysis was used to identify the P9 domain as important for circularization, showing that swapping sequences can restore pairing to improve the circularization of poor circularizers. We also determined the diversity of sequence requirements for the exon 1 and exon 2 (E1 and E2) domains of different group I introns and engineered a S1 tag within the domains for positive purification of circular RNAs. In addition, this flexibility in E1 and E2 enables substitution with less immunostimulatory sequences to enhance protein production. Our work deepens the understanding of the properties of group I introns, expands the panel of introns that can be used, and improves the manufacturing process to generate circular RNAs for vaccines and therapeutics.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

