Atmospheric methane consumption in arid ecosystems acts as a reverse chimney and is accelerated by plant-methanotroph biomes
- PMID: 40037293
- PMCID: PMC11931723
- DOI: 10.1093/ismejo/wraf026
Atmospheric methane consumption in arid ecosystems acts as a reverse chimney and is accelerated by plant-methanotroph biomes
Abstract
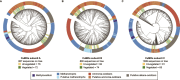
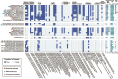
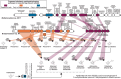
Drylands cover one-third of the Earth's surface and are one of the largest terrestrial sinks for methane. Understanding the structure-function interplay between members of arid biomes can provide critical insights into mechanisms of resilience toward anthropogenic and climate-change-driven environmental stressors-water scarcity, heatwaves, and increased atmospheric greenhouse gases. This study integrates in situ measurements with culture-independent and enrichment-based investigations of methane-consuming microbiomes inhabiting soil in the Anza-Borrego Desert, a model arid ecosystem in Southern California, United States. The atmospheric methane consumption ranged between 2.26 and 12.73 μmol m2 h-1, peaking during the daytime at vegetated sites. Metagenomic studies revealed similar soil-microbiome compositions at vegetated and unvegetated sites, with Methylocaldum being the major methanotrophic clade. Eighty-four metagenome-assembled genomes were recovered, six represented by methanotrophic bacteria (three Methylocaldum, two Methylobacter, and uncultivated Methylococcaceae). The prevalence of copper-containing methane monooxygenases in metagenomic datasets suggests a diverse potential for methane oxidation in canonical methanotrophs and uncultivated Gammaproteobacteria. Five pure cultures of methanotrophic bacteria were obtained, including four Methylocaldum. Genomic analysis of Methylocaldum isolates and metagenome-assembled genomes revealed the presence of multiple stand-alone methane monooxygenase subunit C paralogs, which may have functions beyond methane oxidation. Furthermore, these methanotrophs have genetic signatures typically linked to symbiotic interactions with plants, including tryptophan synthesis and indole-3-acetic acid production. Based on in situ fluxes and soil microbiome compositions, we propose the existence of arid-soil reverse chimneys, an empowered methane sink represented by yet-to-be-defined cooperation between desert vegetation and methane-consuming microbiomes.
Keywords: Methylocaldum; and methane cycle; arid soil; desert microbiomes; methanotrophs; plants.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Dean JF, Middelburg JJ, Röckmann T et al. Methane feedbacks to the global climate system in a warmer world. Rev Geophys 2018;56:207–50. 10.1002/2017RG000559 - DOI
-
- Pachauri RK, Allen MR, Barros VR et al. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Geneva, Switzerland: IPCC, 2014, pp. 151.
-
- Reay DS, Smith P, Christensen TR et al. Methane and global environmental change. Annu Rev Environ Resour 2018;43:165–92. 10.1146/annurev-environ-102017-030154 - DOI
-
- Myhre G, Shindell D, Bréon F-M et al. Anthropogenic and natural radiative forcing. In: Stocker TF, Qin D, Plattner G-K et al. (eds.), Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press, 2013.

