Travel-associated international spread of Oropouche virus beyond the Amazon
- PMID: 40037296
- PMCID: PMC11955161
- DOI: 10.1093/jtm/taaf018
Travel-associated international spread of Oropouche virus beyond the Amazon
Abstract
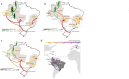
Oropouche virus (OROV), first detected in Trinidad and Tobago in 1955, was historically confined to the Brazilian Amazon Basin. However, since late 2022, an increasing number of OROV cases have been reported across various regions of Brazil as well as in urban centers in Bolivia, Ecuador, Guyana, Colombia, Cuba, Panama, and Peru. In collaboration with Central Public Health Laboratories across Brazil, we integrated epidemiological metadata with genomic analyses from recent cases, generating 133 whole-genome sequences covering the virus's three genomic segments (L, M, and S). These include the first genomes from regions outside the Amazon and from the first recorded fatal cases. Phylogenetic analyses show that the 2024 OROV genomes form a monophyletic group with sequences from the Amazon Basin sampled since 2022, revealing a rapid north-to-south viral movement into historically non-endemic areas. We identified 21 reassortment events, though it remains unclear whether these genomic changes have facilitated viral adaptation to local ecological conditions or contributed to phenotypic traits of public health significance. Our findings demonstrate how OROV has evolved through reassortment and spread rapidly across multiple states in Brazil, leading to the largest outbreak ever recorded outside the Amazon and the first confirmed fatalities. Additionally, by analysing travel-related cases, we provide the first insights into the international spread of OROV beyond Brazil, further highlighting the role of human mobility in its dissemination. The virus's recent rapid geographic expansion and the emergence of severe cases emphasize the urgent need for enhanced surveillance across the Americas. In the absence of significant human population changes over the past two years, factors such as viral adaptation, deforestation, and climate shifts-either individually or in combination-may have facilitated the spread of OROV beyond the Amazon Basin through both local and travel-associated transmission.
Keywords: Amazon basin; Brazil; Orov; genomic surveillance.
© The Author(s) 2025. Published by Oxford University Press on behalf of International Society of Travel Medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- International Committee on Taxonomy of Viruses (ICTV) . Available at: https://ictv.global/taxonomy/. (24 July 2024 date last accessed).
-
- Elliott RM. Orthobunyaviruses: Recent genetic and structural insights. Nat Rev Microbiol 2014; 12:673–85. - PubMed
-
- Anderson CR, Spence L, Downs WG, Aitken TH. Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg 1961; 10:574–8. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- Rede Unificada de Análises Integradas de Arbovírus de Minas Gerais
- MRF-RG-ICCH-2022-100069/MRF_/MRF_/United Kingdom
- NNF24OC0094346/Novo Nordisk Foundation
- GNT2017197/National Health and Medical Research Council
- HTH 017/Rockefeller Foundation
- Health Emergency Preparedness and Response Umbrella Program
- United World Antiviral Research Network
- U01 AI151698/NH/NIH HHS/United States
- RED-00234-23/Fundação de Amparo à Pesquisa do Estado de Minas Gerais
- TF0B8412/World Bank Group
- BIP-00123-23/FAPEMIG
- WT_/Wellcome Trust/United Kingdom
- U01 AI151698/AI/NIAID NIH HHS/United States
- Innovation and Technology Commission, Hong Kong Special Administrative Region, China
- 101046041/European Union's Horizon Europe Research and Innovation Programme
- United World Arbovirus Research Network
- 153597/2024-0/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- CRP/BRA20-03/CRP-ICGEB RESEARCH
- 228186/Z/23/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

