A metabolic synthetic lethality of phosphoinositide 3-kinase-driven cancer
- PMID: 40038309
- PMCID: PMC11880427
- DOI: 10.1038/s41467-025-57225-7
A metabolic synthetic lethality of phosphoinositide 3-kinase-driven cancer
Abstract
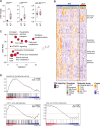
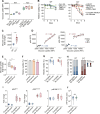
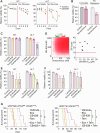
The deregulated activation of the phosphoinositide 3-kinase (PI3K) pathway is a hallmark of aggressive tumors with metabolic plasticity, eliciting their adaptation to the microenvironment and resistance to chemotherapy. A significant gap lies between the biological features of PI3K-driven tumors and the specific targeting of their vulnerabilities. Here, we explore the metabolic liabilities of PI3K-altered T-cell acute lymphoblastic leukemia (T-ALL), an aggressive hematological cancer with dismal outcomes. We report a metabolic crosstalk linking glutaminolysis and glycolysis driven by PI3K signaling alterations. Pharmaceutical inhibition of mTOR reveals the singular plasticity of PI3K-altered cells toward the mobilization of glutamine as a salvage pathway to ensure their survival. Subsequently, the combination of glutamine degradation and mTOR inhibition demonstrates robust cytotoxicity in PI3K-driven solid and hematological tumors in pre-clinical and clinical settings. We propose a novel therapeutic strategy to circumvent metabolic adaptation and efficiently target PI3K-driven cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Vivanco, I. & Sawyers, C. L. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat Rev Cancer. Nat. Publ. Group2, 489–501 (2002). - PubMed
-
- Asnafi, V. et al. Analysis of TCR, pT alpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood101, 2693–2703 (2003). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

