E3 ubiquitin ligase RNF128 promotes Lys63-linked polyubiquitination on SRB1 in macrophages and aggravates atherosclerosis
- PMID: 40038329
- PMCID: PMC11880400
- DOI: 10.1038/s41467-025-57404-6
E3 ubiquitin ligase RNF128 promotes Lys63-linked polyubiquitination on SRB1 in macrophages and aggravates atherosclerosis
Abstract
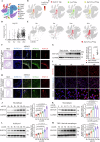
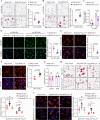
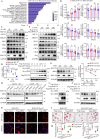
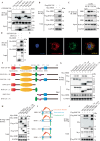
Macrophage-derived foam cell formation is the hallmark of atherosclerotic plaques prominently attributed to excessive lipid uptake and metabolic disorders. As a classic membrane-localized ubiquitin ligase, the role of RNF128 in atherosclerosis remains unknown. We discover that RNF128 is specifically expressed in macrophages of the lipid core based on single-cell RNA sequencing data and persistent hyperlipidemia induces the high expression of RNF128 in macrophages. RNF128 ablation in macrophages ameliorates atherosclerosis in both male and female mice under the background of ApoE and LDLR deficiency. Mechanistically, RNF128 directly binds to scavenger receptor B1 (SRB1), preventing its degradation through the lysosomal system and promoting oxidized low-density lipoprotein (oxLDL)-induced foam cell formation and inflammatory response in macrophages. In addition, RNF128 catalyzes Lys63-linked polyubiquitination on the cytoplasmic C-terminus of the SRB1 at lysine 478, which promotes the endosome SRB1 recycling to the cell membrane with the assistance of Rab11, instead of entering the lysosome for degradation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Libby, P. The changing landscape of atherosclerosis. Nature592, 524–533 (2021). - PubMed
-
- Wang, J. K. et al. Ablation of plasma prekallikrein decreases low-density lipoprotein cholesterol by stabilizing low-density lipoprotein receptor and protects against atherosclerosis. Circulation145, 675–687 (2022). - PubMed
-
- Robinson, J. G. & Gidding, S. S. Curing atherosclerosis should be the next major cardiovascular prevention goal. J Am. Coll. Cardiol.63, 2779–2785 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

