Determinants of response and molecular dynamics in HER2+ER+ breast cancers from the NA-PHER2 trial receiving HER2-targeted and endocrine therapies
- PMID: 40038334
- PMCID: PMC11880565
- DOI: 10.1038/s41467-025-57293-9
Determinants of response and molecular dynamics in HER2+ER+ breast cancers from the NA-PHER2 trial receiving HER2-targeted and endocrine therapies
Abstract
Improved outcomes in HER2+ female breast cancer have resulted from chemotherapy and anti-HER2 therapies. However, HER2+ER+ cancers exhibit lower response rates. The phase 2 NA-PHER2 trial (NCT02530424) investigated chemo-free preoperative HER2 blockade (trastuzumab + pertuzumab) and CDK4/6 inhibition (palbociclib) with or without endocrine therapy (fulvestrant) in HER2+ER+ breast cancer. Clinical endpoints (i.e. Ki67 dynamics and pathological complete response) were previously reported. Here we report on the biomarker analysis, secondary objective of the study. Through RNA sequencing and tumour infiltrating lymphocytes (TIL) assessment in serial biopsies, we identified biomarkers predictive of pCR or Day14 Ki67 response and unveiled treatment-induced molecular changes. High immune infiltration and low ER signalling correlated with pCR, while TP53 mutations associated with high Day14 Ki67. Stratification based on Ki67 at Day14 and at surgery defined three response groups (Ki67 HighHigh, LowHigh, LowLow), with divergent tumour and stroma expression dynamics. The HighHigh group showed dysfunctional immune infiltration and overexpression of therapeutic targets like PAK4 at baseline. The LowLow group exhibited a Luminal A phenotype by the end of treatment. This study expands our understanding of drivers and dynamics of HER2+ER+ tumour response, towards treatment tailoring.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.V. received grants/research supports from: Roche/Genentech, Ventana Medical Systems, Dako/Agilent Technologies; honoraria or consultation feesfrom: Ventana, Dako/Agilent, Roche, MSD Oncology, AstraZeneca, Daiichi Sankyo, Pfizer, Eli Lilly. All the authors declare no conflicts of interest. L.P. has received consulting fees and honoraria for advisory board participation from Pfizer, Astra Zeneca, Merck, Novartis, Bristol-Myers Squibb, Stemline-Menarini, GlaxoSmithKline, Genentech/Roche, Personalis, Daiichi, Natera, Exact Sciences and institutional research funding from Seagen, GlaxoSmithKline, AstraZeneca, Merck, Pfizer and Bristol Myers Squibb. M.Co. received an institutional research grant from Roche. G.B. reports personal fees for consultancy/honorarium/advisory role: Lilly, Novartis, Pfizer, Roche, AstraZeneca, Amgen, MSD, Chugai, Sanofi, Daiichi Sankyo, EISAI, Gilead, Menarini/stemline, Exact Science, Seagen, Agendia; institutional fee for research grant from Gilead. L.G. reports grants from Roche, Breast Cancer Research Foundation (BCRF), and Pfizer during the conduct of the study; personal fees from AstraZeneca, Ely Lilly and Company, Roche, Pfizer, Seattle Genentics, Artemida Pharma, Synaffix, Menarini Ricerche, and Biomedical Insights; grants and personal fees from Zymeworks and Revolution Medicine; other support from METIS Precision Medicine, Amgen, and QU Biologics outside the submitted work; in addition, L.G. has a patent for EU N. 12195182 issued and a patent for EU N.12196177.5 issued.
Figures
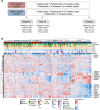






References
-
- Loibl, S. & Gianni, L. HER2-positive breast cancer. Lancet389, 2415–2429 (2017). - PubMed
-
- Bianchini, G. et al. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann. Oncol.26, 2429–2436 (2015). - PubMed
-
- Gianni, L. et al. Neoadjuvant and adjuvant trastuzumab in patients with HER2-positive locally advanced breast cancer (NOAH): Follow-up of a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet Oncol.15, 640–647 (2014). - PubMed
-
- Gianni, L. et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): a multicentre, open-label, phase 2 randomised trial. Lancet Oncol.17, 791–800 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

