Follicular regulatory T cells restrain kidney allograft rejection in mice by suppressing alloreactive B cells
- PMID: 40038336
- PMCID: PMC11880397
- DOI: 10.1038/s41467-025-57468-4
Follicular regulatory T cells restrain kidney allograft rejection in mice by suppressing alloreactive B cells
Abstract
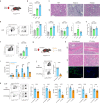
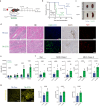
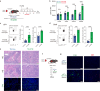
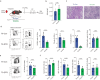
Pathogenic antibodies produced by alloreactive B cells mediate antibody-mediated rejection after kidney transplantation, but the mechanisms remain poorly understood. Follicular regulatory T (Tfr) cells modulate follicular helper T cell-mediated B cell responses, but the functions of Tfr in controlling alloreactive antibody are unknown. Here we study the developmental signals and functions of Tfr cells in mouse allogeneic kidney transplantation models, and show that costimulatory blockade alters the development of Tfr cells disproportionately by decreasing germinal center (GC)-like Tfr cells but increasing follicular-like Tfr cells. Functionally, global Tfr cell deletion results in accelerated graft rejection and increases in donor-specific B cells in both draining lymph nodes and kidney allografts. Mechanistically, Tfr cell deletion increases GC B cell expression of pro-inflammatory cytokines such as IL-15, while neutralization of IL-15 compensates for the loss of Tfr cells and prolongs the survival of mice receiving kidney transplants. Together our preclinical mouse data demonstrate how Tfr restrains kidney allograft rejection by limiting alloreactive B cell responses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- P01 AI056299/AI/NIAID NIH HHS/United States
- P01 AI175397/AI/NIAID NIH HHS/United States
- P01AI056299/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI153124/AI/NIAID NIH HHS/United States
- R35HL166640/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

