Enhanced hepatitis E virus infection of polarised hepatocytes in vitro
- PMID: 40038434
- PMCID: PMC11880378
- DOI: 10.1038/s41598-025-92164-9
Enhanced hepatitis E virus infection of polarised hepatocytes in vitro
Abstract
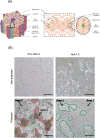
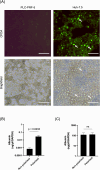
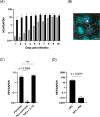
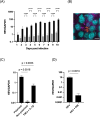
Hepatitis E virus (HEV) is a leading cause of acute viral hepatitis worldwide, and the only zoonotic hepatitis virus. HEV genotype 3 (HEV3) is associated with a range of clinical presentations including chronic infection in immunocompromised individuals in developed nations as well as sporadic cases of autochthonous HEV3 in Europe. Current in vitro models support low levels of HEV infection, hampering our understanding of viral pathogenesis and development of therapeutics. We developed modified culture methods for two widely used hepatoma cell lines, PLC-PRF-5 and Huh-7.5, and evaluated HEV infection. Simple epithelial-like polarity and differentiation formed in PLC-PRF-5 cells, evidenced by localisation of tight junction proteins occludin and zona-occludin 1 to intercellular junctions, and increased albumin production. Complex hepatocyte-like polarity was observed in Huh-7.5 cells, with tight junction proteins localised to shared internal bile canaliculi-like structures and retention of the fluorescent molecule, 5(6)-Carboxyfluorescein diacetate. Cells were infected with genotype 3 HEV, and enhanced infection and replication of HEV was observed using RT-qPCR and immunofluorescent labelling of HEV ORF2 and dsRNA. We describe robust, accessible models for HEV infection in vitro. These models will allow studies to further our understanding of this emerging zoonotic pathogen and develop therapeutic interventions.
Keywords: Hepatitis virus; Hepeviridae; In vitro model; Polarity; Zoonoses.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Izopet, J. et al. Hepatitis E virus infections in Europe. J. Clin. Virol.120, 20–26. 10.1016/j.jcv.2019.09.004 (2019). - PubMed
-
- WHO. Global Hepatitis Report. Report No. 978-92-4-156545-5 (Geneva, 2017).
-
- Purdy, M. A. et al. ICTV virus taxonomy profile: hepeviridae 2022. J. Gen. Virol.10310.1099/jgv.0.001778 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

