Transcriptomic profiling identifies ferroptosis and NF-κB signaling involved in α-dimorphecolic acid regulation of microglial inflammation
- PMID: 40038710
- PMCID: PMC11877847
- DOI: 10.1186/s12967-025-06296-7
Transcriptomic profiling identifies ferroptosis and NF-κB signaling involved in α-dimorphecolic acid regulation of microglial inflammation
Abstract
Background: Microglia-evoked neuroinflammation contributes to neurodegenerative diseases such as multiple sclerosis (MS). Metabolic reprogramming, including changes in polyunsaturated fatty acids (PUFAs), plays a critical role in MS pathophysiology. Previous studies identified reduced plasma α-dimorphecolic acid (α-DIPA), a linoleic acid derivative, in MS patients. This study investigated the anti-inflammatory effects of α-DIPA on microglia and the underlying pathways.
Methods: Lipopolysaccharide (LPS)-induced BV-2 microglial inflammation was used as an in vitro model. α-DIPA effects were assessed via ELISA for nitric oxide (NO) release, flow cytometry was used to examine cell proliferation, activation and polarization, and transcriptomic analysis was applied to identify key signaling pathways regulated by α-DIPA.
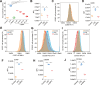
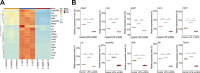
Results: ELISA results showed that exogenous α-DIPA treatment significantly inhibited LPS-induced NO release from BV-2 cells in a concentration-dependent manner. Moreover, flow cytometry analysis suggested that 40 µM α-DIPA treatment significantly repressed LPS-induced BV-2 cell proliferation, activation, as well as M1 and M2 type polarization. Furthermore, transcriptome analysis revealed that exogenous α-DIPA extensively and drastically decreased the transcriptional level of numerous genes that are involved in the regulation of inflammatory responses, for instance, proinflammatory genes such as Tnf and Ccl3 related to IL-17 and TNF-α signaling. In addition, we also observed that the expression of multiple genes in NF-κB signaling were also inhibited greatly by α-DIPA, such as Nfkb2 and Nfkbia. Notably, α-DIPA robustly suppressed LPS-induced mRNA expression of abundant genes participating in the ferroptosis pathway, including Acsl4, Slc7a11, Me1, and Hmox1. Interestingly, the expressions of multiple ferroptosis-related genes were regulated specifically by α-DIPA but not LPS, such as Acsl5, Acsl6, Alox5, Cars, Dpp3, Dpp10, Slc2a5, and Slc7a1.
Conclusion: α-DIPA inhibits microglial inflammation likely through regulating the pathways of the ferroptosis and NF-κB signaling. These results provided preliminary evidence for α-DIPA as a potential therapeutic candidate for neurodegenerative diseases like MS.
Keywords: Ferroptosis; Metabolomics; Microglial inflammation; Multiple sclerosis; NF-κB signaling; Transcriptomics; α-dimorphecolic acid.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The studies involving human participants were reviewed and approved by the Ethics Committee of Lishui Second People’s Hospital (approval number 20171116-3), and the Ethics Committee of Affiliated Hospital of Zunyi Medical University (approval number KLL-2022-305). All subjects provided their written informed consents to participate in this study and for future publication. Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
- QN25H310018/Zhejiang Provincial Natural Science Foundation Youth Science Fund Project
- QianKeHe Basics-ZK [2023] General 583/Basic Research Program of Guizhou Province
- QianKeHe Basics-ZK [2023] General 326/Basic Research Program of Guizhou Province
- Zunshi Kehe HZ Zi (2023) No.259/Basic Research Project of Zunyi City
- WKZX2022JG0105/Project of Development Center for Medical Science & Technology, National Health Commission of the PRC
LinkOut - more resources
Full Text Sources
Miscellaneous

