Motor protein KIF13B orchestrates hepatic metabolism to prevent metabolic dysfunction-associated fatty liver disease
- PMID: 40038775
- PMCID: PMC11877712
- DOI: 10.1186/s40779-025-00594-3
Motor protein KIF13B orchestrates hepatic metabolism to prevent metabolic dysfunction-associated fatty liver disease
Abstract
Background: Kinesin family member 13B (KIF13B), a crucial motor protein, exerts multiple cellular biological functions. However, the implication of KIF13B in metabolic dysfunction-associated fatty liver disease (MAFLD) has not been explored yet. This study aimed to investigate KIF13B's role and underlying mechanism in MAFLD and proposes it as a potential pharmacological target.
Methods: We assessed KIF13B expression in MAFLD patients and rodent models. The roles of Kif13b in lipid metabolism and MAFLD were investigated using whole-body Kif13b knockout mice, hepatocyte-specific Kif13b-deficient mice and hamsters exposed to different diets. The underlying mechanisms by which Kif13b governed hepatic lipid homeostasis and MAFLD progression were explored in vitro. Finally, the Kif13b's impact on atherosclerotic development was studied in the context of MAFLD.
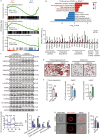
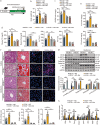
Results: KIF13B expression was reduced in patients and murine models with MAFLD. Rodents with global or liver-specific knockout of the Kif13b gene exhibit spontaneous hepatic steatosis, which is further exacerbated by different overnutrition diets. Overexpression of human KIF13B by lentivirus effectively prevented metabolic dysfunction-associated steatohepatitis (MASH) in methionine-choline-deficient diet (MCD)-fed mice. Furthermore, Kif13b deficiency accelerates atherosclerosis in the context of MAFLD. Mechanistically, Kif13b depletion increases hepatic lipid synthesis and impairs mitochondrial oxidative phosphorylation. Further screening reveals that Kif13b interacts with AMP-activated catalytic subunit alpha 1 (AMPKα1) to regulate the phosphorylation of AMPKα1, governing mitochondrial homeostasis and suppressing sterol regulatory element binding protein 1 (Srebp1)-mediated de novo lipogenesis in the liver.
Conclusion: This work establishes a causal relationship between KIF13B deficiency and MAFLD, emphasizing KIF13B as a potential therapeutic target for treating MAFLD.
Keywords: AMP-activated catalytic subunit alpha 1; Kinesin family member 13B; Lipid metabolism; Metabolic dysfunction-associated fatty liver disease; Mitochondrial homeostasis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: The collection of human liver samples was carried out in accordance with ethical approval from the Medical Ethics Committee of Shengjing Hospital at China Medical University (2023PS913K). Approval for all animal experiments was granted by the Laboratory Animal Ethics Committee of Peking University (LA2023460). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Ann Hepatol. 2024;29(1):101133. - PubMed
-
- Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70(1):151–71. - PubMed
-
- Zhang HJ, He J, Pan LL, Ma ZM, Han CK, Chen CS, et al. Effects of moderate and vigorous exercise on nonalcoholic fatty liver disease: a randomized clinical trial. JAMA Intern Med. 2016;176(8):1074–82. - PubMed
-
- Fromenty B, Roden M. Mitochondrial alterations in fatty liver diseases. J Hepatol. 2023;78(2):415–29. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

