Genomic insights into the plasmidome of non-tuberculous mycobacteria
- PMID: 40038805
- PMCID: PMC11877719
- DOI: 10.1186/s13073-025-01443-7
Genomic insights into the plasmidome of non-tuberculous mycobacteria
Abstract
Background: Non-tuberculous mycobacteria (NTM) are a diverse group of environmental bacteria that are increasingly associated with human infections and difficult to treat. Plasmids, which might carry resistance and virulence factors, remain largely unexplored in NTM.
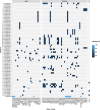
Methods: We used publicly available complete genome sequence data of 328 NTM isolates belonging to 125 species to study gene content, genomic diversity, and clusters of 196 annotated NTM plasmids. Furthermore, we analyzed 3755 draft genome assemblies from over 200 NTM species and 5415 short-read sequence datasets from six clinically relevant NTM species or complexes including M. abscessus, M. avium complex, M. ulcerans complex and M. kansasii complex, for the presence of these plasmids.
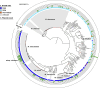
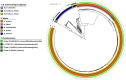
Results: Between one and five plasmids were present in approximately one-third of the complete NTM genomes. The annotated plasmids varied widely in length (most between 10 and 400 kbp) and gene content, with many genes having an unknown function. Predicted gene functions primarily involved plasmid replication, segregation, maintenance, and mobility. Only a few plasmids contained predicted genes that are known to confer resistance to antibiotics commonly used to treat NTM infections. Out of 196 annotated plasmid sequences, 116 could be grouped into 31 clusters of closely related sequences, and about one-third were found across multiple NTM species. Among clinically relevant NTM, the presence of NTM plasmids showed significant variation between species, within (sub)species, and even among strains within (sub)lineages, such as dominant circulating clones of Mycobacterium abscessus.
Conclusions: Our analysis demonstrates that plasmids are a diverse and heterogeneously distributed feature in NTM bacteria. The frequent occurrence of closely related putative plasmid sequences across different NTM species suggests they may play a significant role in NTM evolution through horizontal gene transfer at least in some groups of NTM. However, further in vitro investigations and access to more complete genomes are necessary to validate our findings, elucidate gene functions, identify novel plasmids, and comprehensively assess the role of plasmids in NTM.
Keywords: Antimicrobial resistance; Genomics; Non-tuberculous mycobacteria; Plasmids.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: MD, NW, IB, VD, CU, MM, and SN have no conflicts of interest. FPM is an employee of Basilea Pharmaceutica, Allschwil, Switzerland. FPM contributed to the work presented above outside of this employment. Basilea Pharmaceutica played no role in the conceptualization, conduction, or financing of this study.
Figures




References
-
- Johansen MD, Herrmann JL, Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol. 2020;18:392–407. - PubMed
-
- Van Ingen J, Boeree MJ, Van Soolingen D, Mouton JW. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat. 2012;15:149–61. - PubMed
-
- Dahl VN. et al. Global trends of pulmonary infections with nontuberculous mycobacteria: a systematic review. 2022:S1201–9712. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

