Single and multi-omic characterization of a porcine model of ethanol-induced hepatic fibrosis
- PMID: 40040391
- PMCID: PMC11901410
- DOI: 10.1080/15592294.2025.2471127
Single and multi-omic characterization of a porcine model of ethanol-induced hepatic fibrosis
Abstract
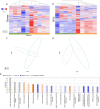
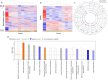
Cirrhosis is a form of end-stage liver disease characterized by extensive hepatic fibrosis and loss of liver parenchyma. It is most commonly the result of long-term alcohol abuse in the United States. Large animal models of cirrhosis, as well as of one of its common long-term sequelae, HCC, are needed to study novel and emerging therapeutic interventions. In the present study, liver fibrosis was induced in the Oncopig cancer model, a large animal HCC model, via intrahepatic, intra-arterial ethanol infusion. Liver sections from five fibrosis induced and five age-matched controls were harvested for RNA-seq (mRNA and lncRNA), small RNA-seq (miRNA), and reduced representation bisulfite sequencing (RRBS; DNA methylation). Single- and multi-omic analysis was performed to investigate the transcriptomic and epigenomic mechanisms associated with fibrosis deposition in this model. A total of 3,439 genes, 70 miRNAs, 452 lncRNAs, and 7,715 methylation regions were found to be differentially regulated through individual single-omic analysis. Pathway analysis indicated differentially expressed genes were associated with collagen synthesis and turnover, hepatic metabolic functions such as ethanol and lipid metabolism, and proliferative and anti-proliferative pathways including PI3K and BAX/BCL signaling pathways. Multi-omic latent variable analysis demonstrated significant concordance with the single-omic analysis. lncRNA's associated with UHRF1BP1L and S1PR1 genes were found to reliably discriminate the two arms of the study. These genes were previously implicated in human cancer development and vasculogenesis, respectively. These findings support the validity and translatability of this model as a useful preclinical tool in the study of alcoholic liver disease and its treatment.
Keywords: Alcoholic liver disease; epigenetics; fibrosis; pig cancer model; translational research.
Conflict of interest statement
Ron Gaba reports grants from United States National Institutes of Health, Guerbet USA LLC, Janssen Research & Development LLC, NeoTherma Oncology, TriSalus Life Sciences, and Sus Clinicals, Inc.; consulting fees from Boston Consulting Group; participation on Sus Clinicals, Inc. Scientific Advisory Board; participation on Fluidx Medical Technology Advisory board; participation on Kaveri University Advisory board; and stock or stock options in Sus Clinicals, Inc. Kyle Schachtschneider and Lawrence Schook report grants from United States National Institutes of Health, United States Department of Agriculture, Society of Interventional Radiology, Guerbet USA LLC, Janssen Research & Development LLC, TriSalus Life Sciences, and Earli Inc. Kyle Schachtschneider and Lawrence Schook are employed by and own equity in Sus Clinicals Inc. Ron Gaba, Lawrence Schook, and Kyle Schachtschneider have a pending patent for Modeling Oncology on Demand, United States Provisional Patent Application No. 62/813,307; Larry and Kyle have a pending patent for Patch for Targeted Delivery of an Oncogenic Cargo to a Tissue, United States Patent Application No. 63/306,449. Both pending patents are licensed to Sus Clinicals..
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
