Low-complexity regions in fungi display functional groups and are depleted in positively charged amino acids
- PMID: 40041205
- PMCID: PMC11878562
- DOI: 10.1093/nargab/lqaf014
Low-complexity regions in fungi display functional groups and are depleted in positively charged amino acids
Abstract
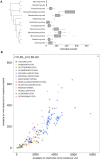
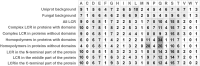
Reports on the diversity and occurrence of low-complexity regions (LCR) in Eukaryota are limited. Some studies have provided a more extensive characterization of LCR proteins in prokaryotes. There is a growing body of knowledge about a plethora of biological functions attributable to LCRs. However, it is hard to determine to what extent observed phenomena apply to fungi since most studies of fungal LCRs were limited to model yeasts. To fill this gap, we performed a survey of LCRs in proteins across all fungal tree of life branches. We show that the abundance of LCRs and the abundance of proteins with LCRs are positively correlated with proteome size. We observed that most LCRs are present in proteins with protein domains but do not overlap with the domain regions. LCRs are associated with many duplicated protein domains. The quantity of particular amino acids in LCRs deviates from the background frequency with a clear over-representation of amino acids with functional groups and a negative charge. Moreover, we discovered that each lineage of fungi favors distinct LCRs expansions. Early diverging fungal lineages differ in LCR abundance and composition pointing at a different evolutionary trajectory of each fungal group.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Comparative functional analysis of proteins containing low-complexity predicted amyloid regions.PeerJ. 2018 Oct 30;6:e5823. doi: 10.7717/peerj.5823. eCollection 2018. PeerJ. 2018. PMID: 30397544 Free PMC article.
-
Low-complexity regions within protein sequences have position-dependent roles.BMC Syst Biol. 2010 Apr 13;4:43. doi: 10.1186/1752-0509-4-43. BMC Syst Biol. 2010. PMID: 20385029 Free PMC article.
-
Dissecting the role of low-complexity regions in the evolution of vertebrate proteins.BMC Evol Biol. 2012 Aug 24;12:155. doi: 10.1186/1471-2148-12-155. BMC Evol Biol. 2012. PMID: 22920595 Free PMC article.
-
Evolutionary Trajectories of Entomopathogenic Fungi ABC Transporters.Adv Genet. 2017;98:117-154. doi: 10.1016/bs.adgen.2017.07.002. Epub 2017 Aug 4. Adv Genet. 2017. PMID: 28942792 Review.
-
Disentangling the complexity of low complexity proteins.Brief Bioinform. 2020 Mar 23;21(2):458-472. doi: 10.1093/bib/bbz007. Brief Bioinform. 2020. PMID: 30698641 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

