Decreased levels and function of dendritic cells in blood and airways predict COVID-19 severity
- PMID: 40041475
- PMCID: PMC11873541
- DOI: 10.1002/cti2.70026
Decreased levels and function of dendritic cells in blood and airways predict COVID-19 severity
Abstract
Objectives: Monocytes and dendritic cells (DCs) are essential players in the immune response to infections, involved in shaping innate and adaptive immunity. However, a complete understanding of their specific roles in respiratory infections, including SARS-CoV-2, remains elusive.
Methods: To investigate the dynamics of monocytes and DCs in blood as well as the upper and lower airways, we sampled 147 patients with varying degree of COVID-19 severity longitudinally during the spring of 2020.
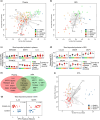
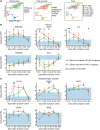
Results: Using flow cytometry, proteomics and in vitro TLR stimulation, we found differences in the distribution and function of monocytes and DCs in patients compared with controls, and importantly, reduced levels of DCs in both blood and airways. In fact, lower frequencies of cDC2s (Lin- HLA-DR+ CD1c+) early after symptom onset predicted subsequent severe disease, and depletion of DC subsets lasted longer in patients with more severe disease. In contrast, severe COVID-19 was associated with increased frequencies of activated monocytes in the lower, but not the upper, airways. Proteomic analysis showed that monocyte and DC-related cytokines in plasma and airways associated with disease severity. During convalescence, cell frequencies and responses to TLR ligands normalised in blood, except for persistently low plasmacytoid DCs.
Conclusion: Our study reveals a distinct pattern of recruitment of monocytes but not DCs to the airways during severe COVID-19. Instead, decreased levels of DCs in both blood and airways were found, possibly contributing to more severe COVID-19. The connection between low blood DCs early in disease course and more severe outcomes provides insight into COVID-19 immunopathology, with possible therapeutic implications.
Keywords: COVID‐19; SARS‐CoV‐2; dendritic cells; monocytes; respiratory immunology.
© 2025 The Author(s). Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
AS‐S is a paid consultant to Astra‐Zeneca on COVID‐19 clinical trials not related to this study.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
